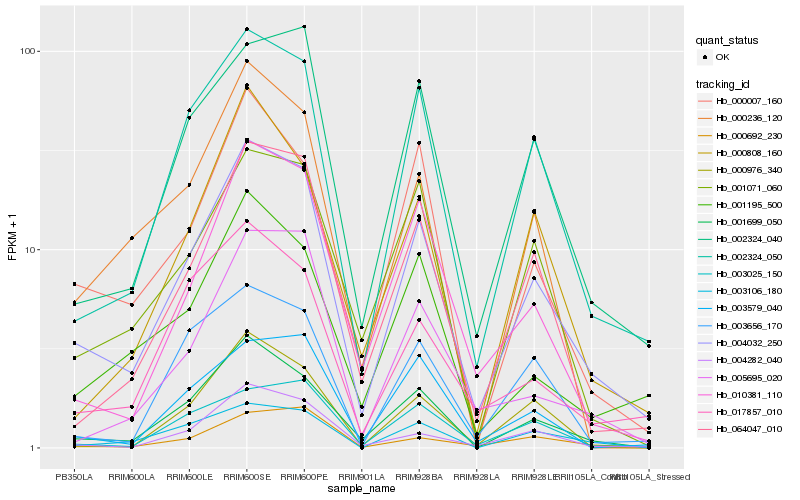
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004032_250 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002324_050 |
0.1029796667 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 3 |
Hb_001699_050 |
0.1128038971 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s11410g [Populus trichocarpa] |
| 4 |
Hb_002324_040 |
0.1214013917 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 5 |
Hb_064047_010 |
0.1236633815 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004282_040 |
0.1333461136 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_010381_110 |
0.1372290006 |
- |
- |
PRA1 family protein [Theobroma cacao] |
| 8 |
Hb_003025_150 |
0.1469057151 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001071_060 |
0.147990341 |
- |
- |
Trehalose phosphatase/synthase 5 isoform 1 [Theobroma cacao] |
| 10 |
Hb_000007_160 |
0.1494260855 |
- |
- |
hypothetical protein POPTR_0008s10970g [Populus trichocarpa] |
| 11 |
Hb_000236_120 |
0.1504540328 |
- |
- |
- |
| 12 |
Hb_001195_500 |
0.1511635446 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 13 |
Hb_003106_180 |
0.1511792209 |
- |
- |
Peroxidase 65 precursor, putative [Ricinus communis] |
| 14 |
Hb_003579_040 |
0.1530310649 |
- |
- |
PREDICTED: monoacylglycerol lipase ABHD6 [Jatropha curcas] |
| 15 |
Hb_000692_230 |
0.1535376258 |
- |
- |
phospholipase d delta, putative [Ricinus communis] |
| 16 |
Hb_003656_170 |
0.1547815806 |
- |
- |
PREDICTED: phenylalanine ammonia-lyase-like [Jatropha curcas] |
| 17 |
Hb_000808_160 |
0.155007368 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |
| 18 |
Hb_017857_010 |
0.1563454922 |
- |
- |
- |
| 19 |
Hb_005695_020 |
0.1563767573 |
- |
- |
PREDICTED: low affinity sulfate transporter 3 [Jatropha curcas] |
| 20 |
Hb_000976_340 |
0.1568608822 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |