| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
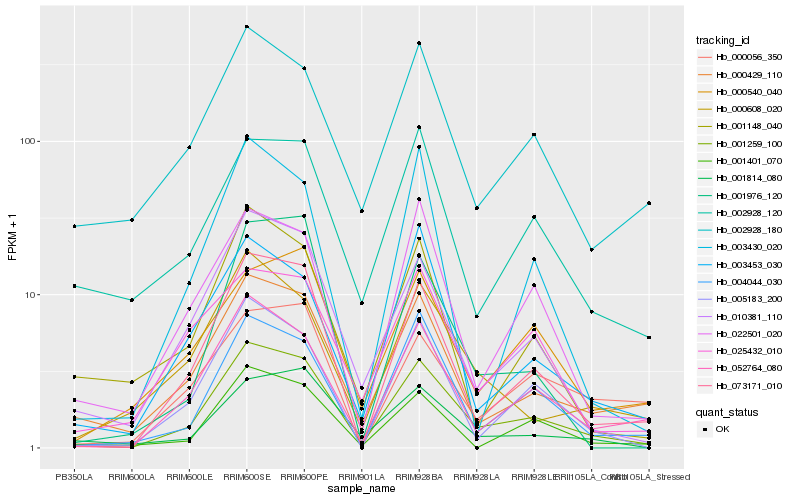
Hb_001259_100 |
0.0 |
- |
- |
chloride channel-like family protein [Populus trichocarpa] |
| 2 |
Hb_073171_010 |
0.1034776095 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 3 |
Hb_052764_080 |
0.120877889 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD5 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005183_200 |
0.1335711642 |
- |
- |
EG964 [Manihot glaziovii] |
| 5 |
Hb_000429_110 |
0.1348758799 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2-like [Populus euphratica] |
| 6 |
Hb_004044_030 |
0.1351189403 |
- |
- |
PREDICTED: uncharacterized protein LOC105639335 [Jatropha curcas] |
| 7 |
Hb_010381_110 |
0.136642165 |
- |
- |
PRA1 family protein [Theobroma cacao] |
| 8 |
Hb_022501_020 |
0.15397713 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 9 |
Hb_001401_070 |
0.1633853552 |
- |
- |
glycogenin, putative [Ricinus communis] |
| 10 |
Hb_003453_030 |
0.1635282046 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 11 |
Hb_002928_180 |
0.1660987478 |
- |
- |
catalase CAT1 [Manihot esculenta] |
| 12 |
Hb_000608_020 |
0.1666719438 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 13 |
Hb_003430_020 |
0.1668940411 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_001814_080 |
0.1671261565 |
- |
- |
PREDICTED: oleosin 1-like [Jatropha curcas] |
| 15 |
Hb_000540_040 |
0.1713394449 |
- |
- |
PREDICTED: chitinase 2 [Amborella trichopoda] |
| 16 |
Hb_025432_010 |
0.1723891277 |
- |
- |
hypothetical protein JCGZ_21627 [Jatropha curcas] |
| 17 |
Hb_001148_040 |
0.1736060566 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 18 |
Hb_000056_350 |
0.1763584414 |
- |
- |
PREDICTED: uncharacterized protein LOC105630437 [Jatropha curcas] |
| 19 |
Hb_001976_120 |
0.176657471 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 20 |
Hb_002928_120 |
0.1770013353 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |