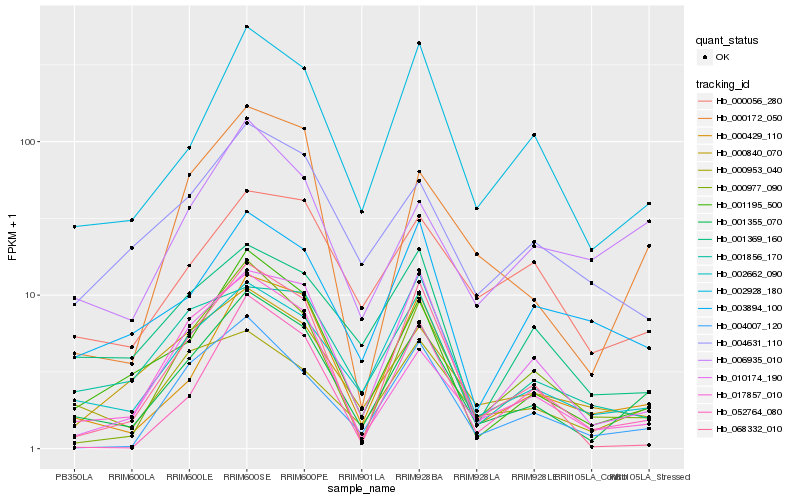
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001355_070 |
0.0 |
- |
- |
P-loop containing nucleoside triphosphate hydrolases superfamily protein, putative isoform 2 [Theobroma cacao] |
| 2 |
Hb_001195_500 |
0.1335252192 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 3 |
Hb_002928_180 |
0.1360320064 |
- |
- |
catalase CAT1 [Manihot esculenta] |
| 4 |
Hb_000429_110 |
0.1393329055 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2-like [Populus euphratica] |
| 5 |
Hb_000172_050 |
0.1526504351 |
- |
- |
PREDICTED: uncharacterized protein LOC105650611 [Jatropha curcas] |
| 6 |
Hb_002662_090 |
0.1534203118 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 7 |
Hb_010174_190 |
0.1545256552 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 8 |
Hb_000977_090 |
0.156464252 |
- |
- |
hypothetical protein POPTR_0010s18550g [Populus trichocarpa] |
| 9 |
Hb_000953_040 |
0.1667458714 |
- |
- |
hypothetical protein OsJ_19135 [Oryza sativa Japonica Group] |
| 10 |
Hb_004007_120 |
0.1683106761 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type isoform X1 [Populus euphratica] |
| 11 |
Hb_006935_010 |
0.17467284 |
- |
- |
PREDICTED: inositol transporter 1 isoform X1 [Gossypium raimondii] |
| 12 |
Hb_000840_070 |
0.176472262 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_017857_010 |
0.1776984607 |
- |
- |
- |
| 14 |
Hb_004631_110 |
0.1818160333 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 15 |
Hb_068332_010 |
0.1824104249 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 16 |
Hb_000056_280 |
0.182895892 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 17 |
Hb_001369_160 |
0.1845331794 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 18 |
Hb_052764_080 |
0.1850266189 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD5 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001856_170 |
0.1855066977 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 20 |
Hb_003894_100 |
0.1860054129 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1-like [Jatropha curcas] |