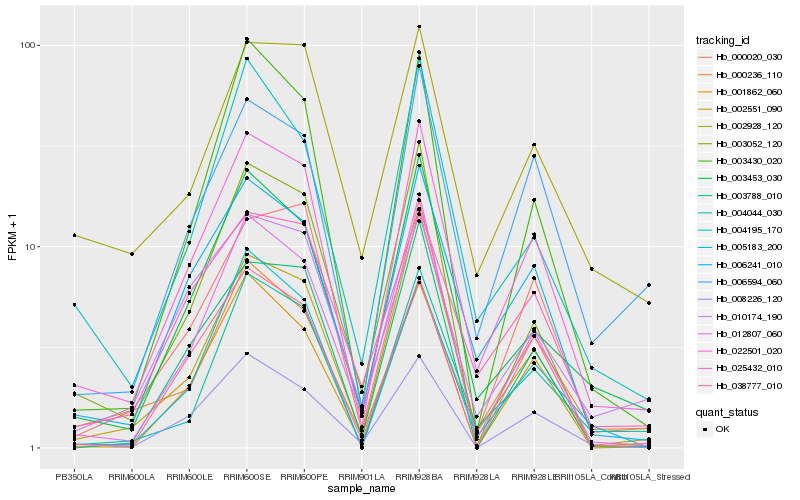
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022501_020 |
0.0 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 2 |
Hb_006241_010 |
0.0822331021 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 3 |
Hb_012807_060 |
0.1084219109 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_003453_030 |
0.1104972278 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 5 |
Hb_002551_090 |
0.1123752771 |
- |
- |
hypothetical protein JCGZ_17224 [Jatropha curcas] |
| 6 |
Hb_004044_030 |
0.1124515949 |
- |
- |
PREDICTED: uncharacterized protein LOC105639335 [Jatropha curcas] |
| 7 |
Hb_006594_060 |
0.115893077 |
transcription factor |
TF Family: HSF |
PREDICTED: heat shock factor protein HSF24 [Jatropha curcas] |
| 8 |
Hb_008226_120 |
0.115908374 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_000020_030 |
0.1164926091 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_025432_010 |
0.118810235 |
- |
- |
hypothetical protein JCGZ_21627 [Jatropha curcas] |
| 11 |
Hb_003430_020 |
0.1228940302 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_004195_170 |
0.1256633826 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_000236_110 |
0.1265049084 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic [Jatropha curcas] |
| 14 |
Hb_010174_190 |
0.128867378 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 15 |
Hb_038777_010 |
0.1289865897 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN2 isoform X1 [Populus euphratica] |
| 16 |
Hb_005183_200 |
0.129627273 |
- |
- |
EG964 [Manihot glaziovii] |
| 17 |
Hb_001862_060 |
0.1339594426 |
- |
- |
PREDICTED: uncharacterized protein LOC105639802 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003052_120 |
0.1388796589 |
- |
- |
hypothetical protein POPTR_0008s10970g [Populus trichocarpa] |
| 19 |
Hb_003788_010 |
0.1391557301 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 20 |
Hb_002928_120 |
0.1398592742 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |