| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
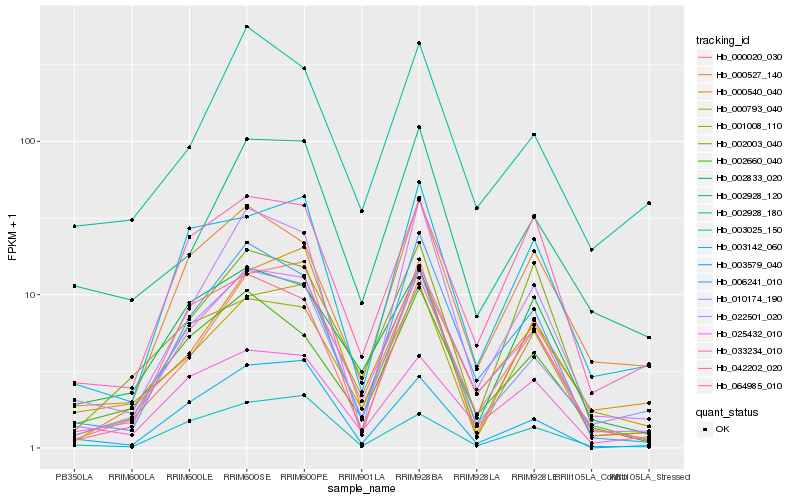
Hb_025432_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_21627 [Jatropha curcas] |
| 2 |
Hb_010174_190 |
0.0938516116 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 3 |
Hb_006241_010 |
0.0987029764 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 4 |
Hb_064985_010 |
0.1139580748 |
- |
- |
Leucine-rich repeat transmembrane protein kinase, putative [Theobroma cacao] |
| 5 |
Hb_001008_110 |
0.1169480021 |
- |
- |
- |
| 6 |
Hb_022501_020 |
0.118810235 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 7 |
Hb_042202_020 |
0.1235954428 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 8 |
Hb_033234_010 |
0.1256970732 |
desease resistance |
Gene Name: LRR_8 |
Disease resistance protein RPP8 [Theobroma cacao] |
| 9 |
Hb_000527_140 |
0.1264799355 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 10 |
Hb_002660_040 |
0.129406193 |
- |
- |
PREDICTED: CASP-like protein 5A1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003142_060 |
0.1336802923 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_000540_040 |
0.1343196998 |
- |
- |
PREDICTED: chitinase 2 [Amborella trichopoda] |
| 13 |
Hb_000020_030 |
0.1348812607 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_002928_120 |
0.135842514 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 15 |
Hb_002928_180 |
0.1370837161 |
- |
- |
catalase CAT1 [Manihot esculenta] |
| 16 |
Hb_000793_040 |
0.1372198138 |
- |
- |
PREDICTED: transcription factor LHW [Jatropha curcas] |
| 17 |
Hb_002833_020 |
0.1417597743 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 18 |
Hb_003579_040 |
0.1423774051 |
- |
- |
PREDICTED: monoacylglycerol lipase ABHD6 [Jatropha curcas] |
| 19 |
Hb_003025_150 |
0.1464867126 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002003_040 |
0.1505265614 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |