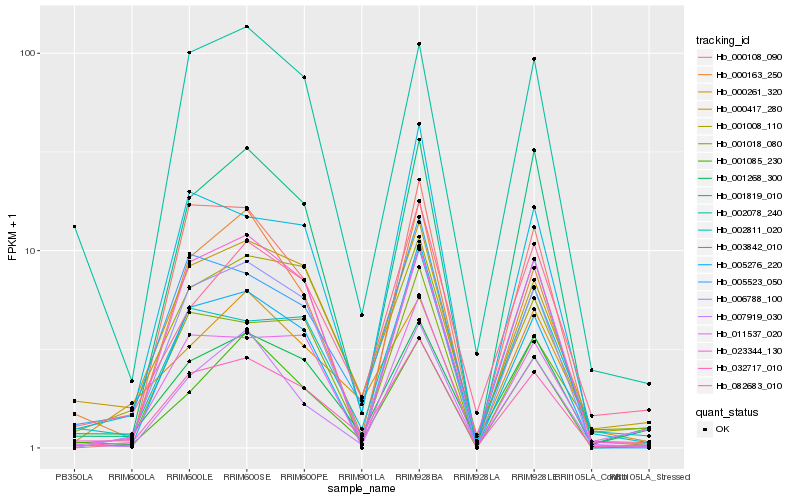
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032717_010 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_006788_100 |
0.0936424342 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000417_280 |
0.0984331709 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005523_050 |
0.1020028998 |
- |
- |
hypothetical protein JCGZ_09175 [Jatropha curcas] |
| 5 |
Hb_001008_110 |
0.1095104782 |
- |
- |
- |
| 6 |
Hb_001268_300 |
0.1169860597 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001819_010 |
0.1186528455 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 8 |
Hb_005276_220 |
0.1186960153 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 9 |
Hb_000163_250 |
0.119011176 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_002811_020 |
0.1281484737 |
- |
- |
Cysteine synthase [Morus notabilis] |
| 11 |
Hb_023344_130 |
0.1287355098 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_011537_020 |
0.1299840358 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13-like [Jatropha curcas] |
| 13 |
Hb_001085_230 |
0.1318794897 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |
| 14 |
Hb_007919_030 |
0.1322036855 |
- |
- |
- |
| 15 |
Hb_082683_010 |
0.1353827779 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_003842_010 |
0.1367828343 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 17 |
Hb_001018_080 |
0.1380869494 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000108_090 |
0.1383636132 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 19 |
Hb_002078_240 |
0.1390882372 |
rubber biosynthesis |
Gene Name: SRPP4 |
PREDICTED: REF/SRPP-like protein At1g67360 [Jatropha curcas] |
| 20 |
Hb_000261_320 |
0.1407751942 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |