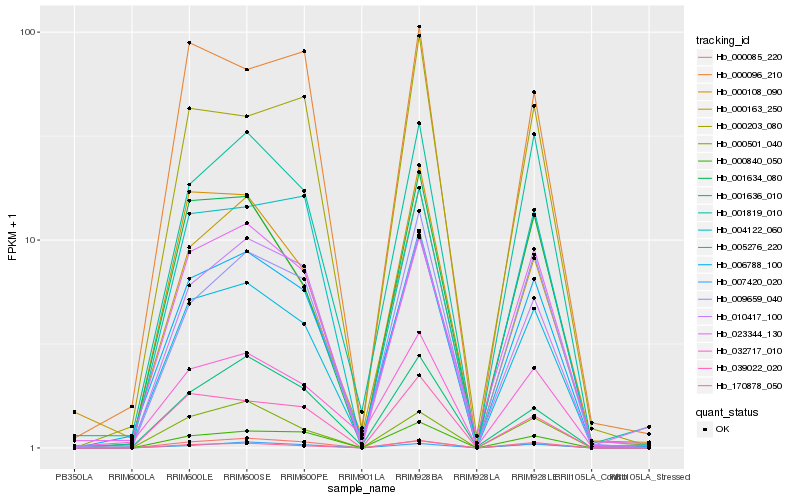
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006788_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001819_010 |
0.0905876044 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 3 |
Hb_023344_130 |
0.0906994363 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_032717_010 |
0.0936424342 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_005276_220 |
0.0989730233 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 6 |
Hb_000840_050 |
0.101072042 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Vitis vinifera] |
| 7 |
Hb_010417_100 |
0.1085677193 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A [Jatropha curcas] |
| 8 |
Hb_000108_090 |
0.1094533044 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 9 |
Hb_009659_040 |
0.1098565969 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER [Populus euphratica] |
| 10 |
Hb_000163_250 |
0.1113850629 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_004122_060 |
0.1146888022 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280-like [Jatropha curcas] |
| 12 |
Hb_001634_080 |
0.1170663672 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_000501_040 |
0.1177330663 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_000085_220 |
0.1192406688 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000203_080 |
0.1192783539 |
- |
- |
Xylem bark cysteine peptidase 3 isoform 1 [Theobroma cacao] |
| 16 |
Hb_039022_020 |
0.1193360569 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 17 |
Hb_007420_020 |
0.1204816764 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 18 |
Hb_000096_210 |
0.1217073831 |
- |
- |
PREDICTED: probable carboxylesterase 7 [Jatropha curcas] |
| 19 |
Hb_001636_010 |
0.1221678312 |
- |
- |
hypothetical protein JCGZ_01264 [Jatropha curcas] |
| 20 |
Hb_170878_050 |
0.1222125916 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |