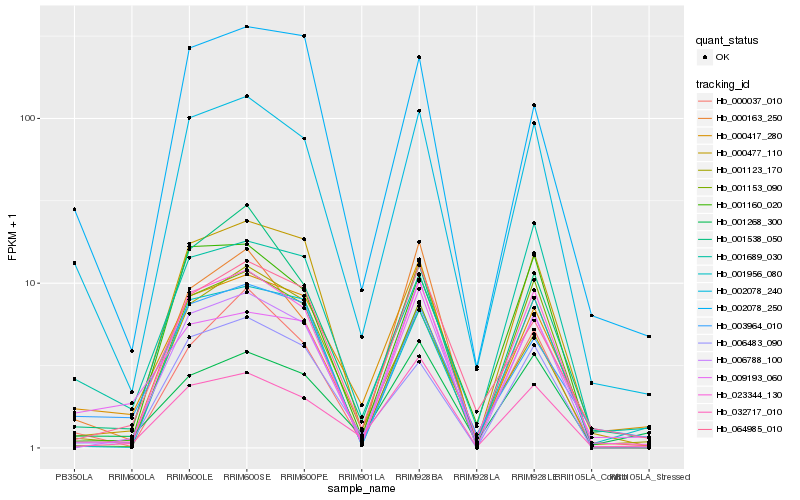
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002078_240 |
0.0 |
rubber biosynthesis |
Gene Name: SRPP4 |
PREDICTED: REF/SRPP-like protein At1g67360 [Jatropha curcas] |
| 2 |
Hb_023344_130 |
0.0985768297 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_000417_280 |
0.1218550322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000037_010 |
0.1241269678 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Jatropha curcas] |
| 5 |
Hb_003964_010 |
0.132075832 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0008s09360g [Populus trichocarpa] |
| 6 |
Hb_000477_110 |
0.1321080657 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 7 |
Hb_001268_300 |
0.1334232396 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001160_020 |
0.13492287 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 9 |
Hb_001153_090 |
0.1382281253 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 10 |
Hb_006483_090 |
0.1384700834 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 11 |
Hb_001123_170 |
0.1384756882 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_032717_010 |
0.1390882372 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_001689_030 |
0.14030498 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 14 |
Hb_009193_060 |
0.1403877136 |
- |
- |
PREDICTED: ureide permease 1-like [Jatropha curcas] |
| 15 |
Hb_000163_250 |
0.1408776707 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_001956_080 |
0.1410231807 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 17 |
Hb_001538_050 |
0.1411689069 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 18 |
Hb_006788_100 |
0.142668805 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002078_250 |
0.143733769 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_064985_010 |
0.1457747389 |
- |
- |
Leucine-rich repeat transmembrane protein kinase, putative [Theobroma cacao] |