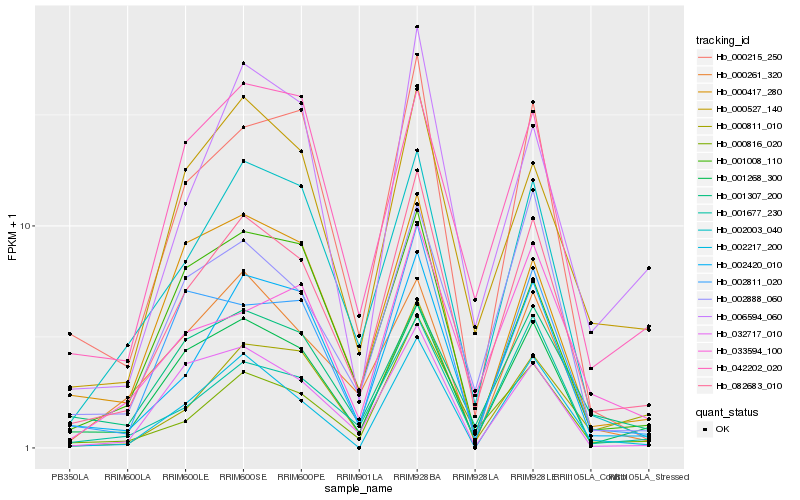
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_082683_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_002420_010 |
0.101979659 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 3 |
Hb_001268_300 |
0.1075489164 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000816_020 |
0.1083010072 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_000527_140 |
0.1197945746 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 6 |
Hb_002003_040 |
0.1214028668 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001677_230 |
0.1271134954 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000215_250 |
0.1281101542 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_000811_010 |
0.1317191687 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF118 [Jatropha curcas] |
| 10 |
Hb_006594_060 |
0.1339206831 |
transcription factor |
TF Family: HSF |
PREDICTED: heat shock factor protein HSF24 [Jatropha curcas] |
| 11 |
Hb_002811_020 |
0.1349025454 |
- |
- |
Cysteine synthase [Morus notabilis] |
| 12 |
Hb_032717_010 |
0.1353827779 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_002217_200 |
0.1373011155 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 14 |
Hb_002888_060 |
0.1378714532 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001008_110 |
0.146662565 |
- |
- |
- |
| 16 |
Hb_042202_020 |
0.1481684619 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 17 |
Hb_000261_320 |
0.1487617383 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_001307_200 |
0.1496796941 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 19 |
Hb_033594_100 |
0.150193107 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_000417_280 |
0.1502486082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |