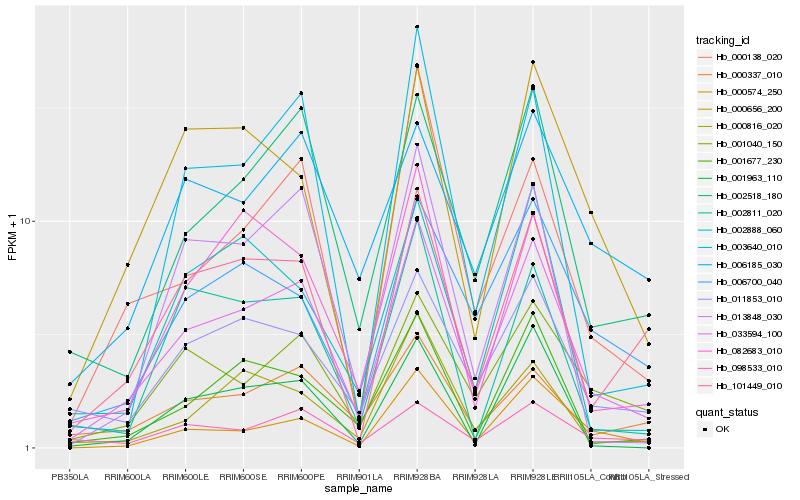
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033594_100 |
0.0 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_002518_180 |
0.1242402201 |
- |
- |
hypothetical protein POPTR_0001s25460g [Populus trichocarpa] |
| 3 |
Hb_011853_010 |
0.1359271959 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 4 |
Hb_001040_150 |
0.1391142312 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001677_230 |
0.1423315644 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_101449_010 |
0.14344567 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 7 |
Hb_082683_010 |
0.150193107 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_006700_040 |
0.1524328472 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 9 |
Hb_000337_010 |
0.1539456273 |
- |
- |
PREDICTED: proteasome activator subunit 4 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000138_020 |
0.1541413102 |
- |
- |
PREDICTED: peroxidase A2-like [Jatropha curcas] |
| 11 |
Hb_000574_250 |
0.154641194 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 12 |
Hb_000816_020 |
0.1550643469 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_013848_030 |
0.1569163379 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 14 |
Hb_003640_010 |
0.1618171235 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 15 |
Hb_000656_200 |
0.1627846521 |
- |
- |
PREDICTED: uncharacterized protein LOC105631373 [Jatropha curcas] |
| 16 |
Hb_001963_110 |
0.1643556777 |
- |
- |
- |
| 17 |
Hb_098533_010 |
0.1644309424 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 18 |
Hb_002811_020 |
0.1662453588 |
- |
- |
Cysteine synthase [Morus notabilis] |
| 19 |
Hb_006185_030 |
0.1665567728 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 20 |
Hb_002888_060 |
0.1679601701 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |