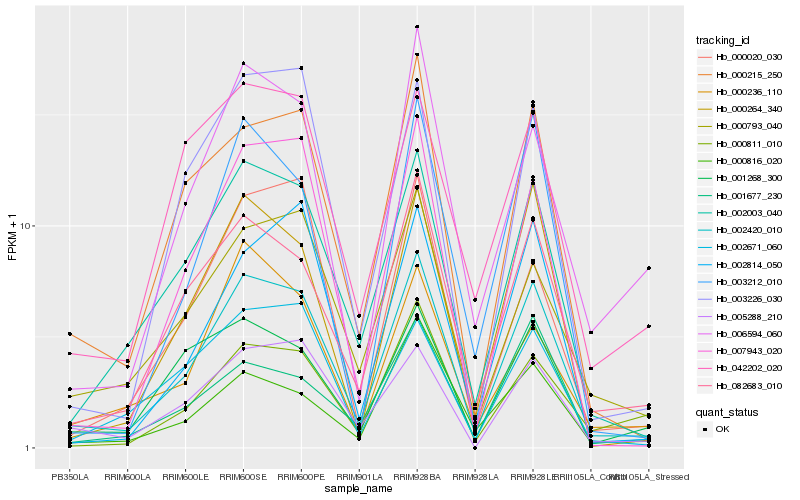
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002420_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 2 |
Hb_002003_040 |
0.0963545033 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 3 |
Hb_082683_010 |
0.101979659 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_000215_250 |
0.1097808833 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_007943_020 |
0.1248537115 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_000264_340 |
0.1252180491 |
- |
- |
PREDICTED: callose synthase 11-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000793_040 |
0.129373938 |
- |
- |
PREDICTED: transcription factor LHW [Jatropha curcas] |
| 8 |
Hb_005288_210 |
0.1309314903 |
- |
- |
hypothetical protein VITISV_017217 [Vitis vinifera] |
| 9 |
Hb_003226_030 |
0.1399642616 |
- |
- |
sucrose synthase 5 [Hevea brasiliensis] |
| 10 |
Hb_006594_060 |
0.140941286 |
transcription factor |
TF Family: HSF |
PREDICTED: heat shock factor protein HSF24 [Jatropha curcas] |
| 11 |
Hb_001268_300 |
0.141126586 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_042202_020 |
0.1441470706 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 13 |
Hb_003212_010 |
0.1441741932 |
- |
- |
sucrose transporter 4 [Hevea brasiliensis] |
| 14 |
Hb_002814_050 |
0.1476321668 |
- |
- |
PREDICTED: sugar carrier protein A [Jatropha curcas] |
| 15 |
Hb_000811_010 |
0.1478065325 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF118 [Jatropha curcas] |
| 16 |
Hb_001677_230 |
0.1480689561 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002671_060 |
0.1507705133 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 18 |
Hb_000816_020 |
0.1511073158 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_000020_030 |
0.1535931531 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_000236_110 |
0.1540131395 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic [Jatropha curcas] |