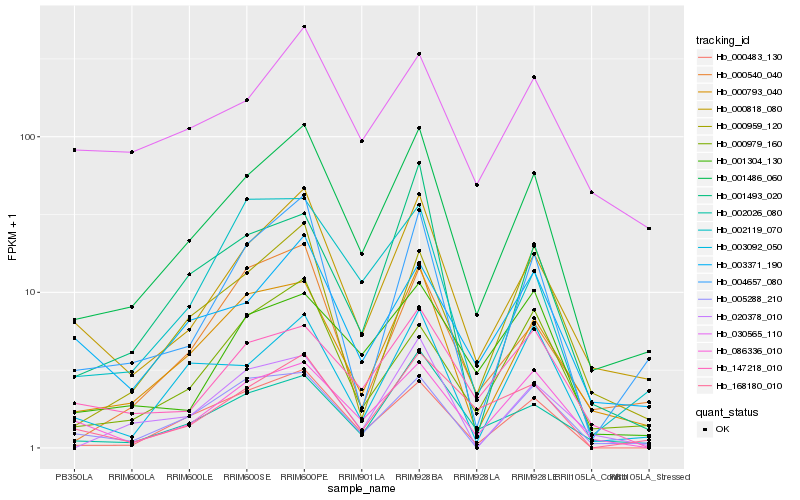
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001486_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632622 [Jatropha curcas] |
| 2 |
Hb_000818_080 |
0.0992144032 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 3 |
Hb_000793_040 |
0.1093194869 |
- |
- |
PREDICTED: transcription factor LHW [Jatropha curcas] |
| 4 |
Hb_002026_080 |
0.1326508069 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 5 |
Hb_000483_130 |
0.1366580004 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 6 |
Hb_086336_010 |
0.1467563619 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 7 |
Hb_004657_080 |
0.1487011641 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 8 |
Hb_168180_010 |
0.149404258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003371_190 |
0.1518215217 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 10 |
Hb_003092_050 |
0.1527622326 |
- |
- |
PREDICTED: putative chloride channel-like protein CLC-g [Jatropha curcas] |
| 11 |
Hb_000979_160 |
0.1529194801 |
- |
- |
hypothetical protein RCOM_0629030 [Ricinus communis] |
| 12 |
Hb_005288_210 |
0.1533283188 |
- |
- |
hypothetical protein VITISV_017217 [Vitis vinifera] |
| 13 |
Hb_002119_070 |
0.1605638883 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_000959_120 |
0.1606982513 |
- |
- |
PREDICTED: protein BRASSINOSTEROID INSENSITIVE 1 [Jatropha curcas] |
| 15 |
Hb_147218_010 |
0.1618216146 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 16 |
Hb_000540_040 |
0.1633771676 |
- |
- |
PREDICTED: chitinase 2 [Amborella trichopoda] |
| 17 |
Hb_030565_110 |
0.1641792233 |
- |
- |
PREDICTED: adenosylhomocysteinase 1 [Jatropha curcas] |
| 18 |
Hb_001493_020 |
0.1642280249 |
- |
- |
leucine-rich repeat receptor-like protein kinase [Manihot esculenta] |
| 19 |
Hb_001304_130 |
0.1654025577 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like transcription factor PAT1 [Jatropha curcas] |
| 20 |
Hb_020378_010 |
0.1669255745 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1 [Jatropha curcas] |