| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
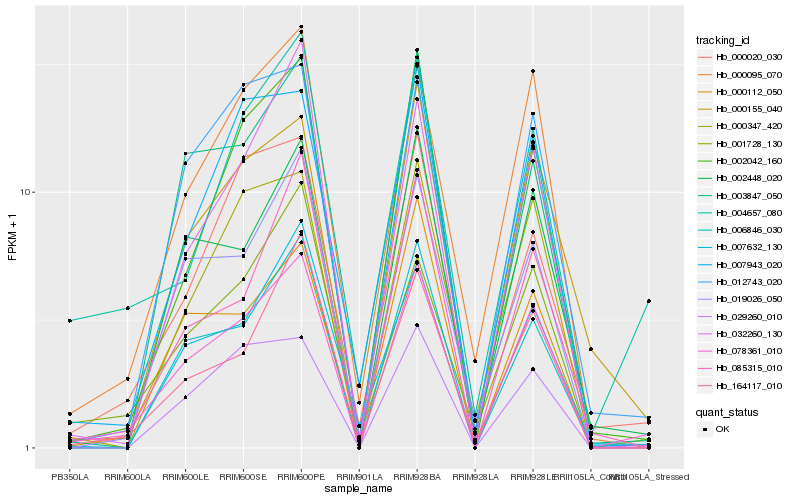
Hb_078361_010 |
0.0 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 2 |
Hb_006846_030 |
0.1235878463 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 3 |
Hb_000095_070 |
0.1279770471 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001728_130 |
0.1322998945 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_012743_020 |
0.1327912786 |
- |
- |
serine/threonine protein phosphatase, putative [Ricinus communis] |
| 6 |
Hb_002042_160 |
0.1336326509 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 7 |
Hb_164117_010 |
0.1342655647 |
- |
- |
hypothetical protein PHAVU_004G0480000g [Phaseolus vulgaris] |
| 8 |
Hb_032260_130 |
0.1353430652 |
- |
- |
hypothetical protein POPTR_0003s13800g [Populus trichocarpa] |
| 9 |
Hb_002448_020 |
0.1376661743 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 10 |
Hb_019026_050 |
0.1389493541 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003847_050 |
0.1400670164 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 12 |
Hb_000155_040 |
0.1423374518 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 13 |
Hb_000020_030 |
0.1436005472 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_007632_130 |
0.1440863491 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_085315_010 |
0.1444755095 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 16 |
Hb_000347_420 |
0.1448335629 |
- |
- |
hypothetical protein JCGZ_24696 [Jatropha curcas] |
| 17 |
Hb_029260_010 |
0.1450635828 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 18 |
Hb_000112_050 |
0.1454219319 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |
| 19 |
Hb_004657_080 |
0.1471259534 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 20 |
Hb_007943_020 |
0.1479872028 |
- |
- |
transcription factor, putative [Ricinus communis] |