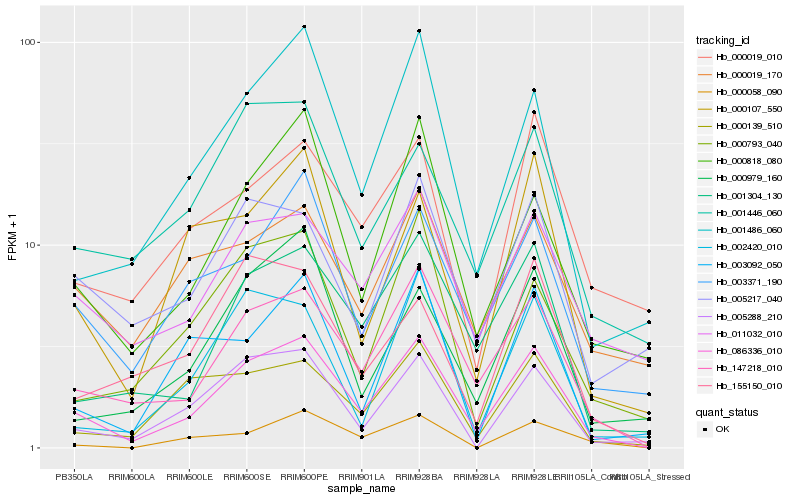
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_086336_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 2 |
Hb_147218_010 |
0.1044035128 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 3 |
Hb_001486_060 |
0.1467563619 |
- |
- |
PREDICTED: uncharacterized protein LOC105632622 [Jatropha curcas] |
| 4 |
Hb_001304_130 |
0.1486328908 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like transcription factor PAT1 [Jatropha curcas] |
| 5 |
Hb_003371_190 |
0.1520536319 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 6 |
Hb_000818_080 |
0.1534749305 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 7 |
Hb_005288_210 |
0.1571533564 |
- |
- |
hypothetical protein VITISV_017217 [Vitis vinifera] |
| 8 |
Hb_011032_010 |
0.1614086187 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 9 |
Hb_000139_510 |
0.1688375147 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000107_550 |
0.1695496523 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 11 |
Hb_000793_040 |
0.1727185267 |
- |
- |
PREDICTED: transcription factor LHW [Jatropha curcas] |
| 12 |
Hb_001446_060 |
0.1744667172 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_005217_040 |
0.1753217057 |
- |
- |
PREDICTED: F-box protein PP2-A12 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002420_010 |
0.176849467 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 15 |
Hb_000979_160 |
0.1772242359 |
- |
- |
hypothetical protein RCOM_0629030 [Ricinus communis] |
| 16 |
Hb_003092_050 |
0.1816205967 |
- |
- |
PREDICTED: putative chloride channel-like protein CLC-g [Jatropha curcas] |
| 17 |
Hb_000019_170 |
0.1817738086 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 18 |
Hb_155150_010 |
0.1830990146 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_000058_090 |
0.1833811411 |
- |
- |
- |
| 20 |
Hb_000019_010 |
0.1835918968 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Jatropha curcas] |