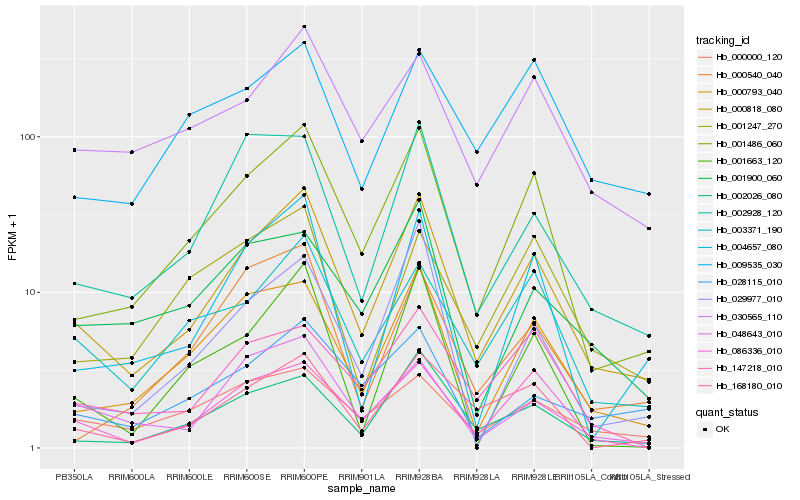
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000818_080 |
0.0 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 2 |
Hb_001486_060 |
0.0992144032 |
- |
- |
PREDICTED: uncharacterized protein LOC105632622 [Jatropha curcas] |
| 3 |
Hb_000793_040 |
0.1374596296 |
- |
- |
PREDICTED: transcription factor LHW [Jatropha curcas] |
| 4 |
Hb_002026_080 |
0.1388151299 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 5 |
Hb_003371_190 |
0.1454511963 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 6 |
Hb_002928_120 |
0.1487117328 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 7 |
Hb_004657_080 |
0.1502102498 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 8 |
Hb_168180_010 |
0.1508706297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_086336_010 |
0.1534749305 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 10 |
Hb_030565_110 |
0.155033873 |
- |
- |
PREDICTED: adenosylhomocysteinase 1 [Jatropha curcas] |
| 11 |
Hb_048643_010 |
0.1569512081 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 12 |
Hb_028115_010 |
0.1628813717 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000000_120 |
0.1648181424 |
- |
- |
hypothetical protein EUGRSUZ_A004731, partial [Eucalyptus grandis] |
| 14 |
Hb_147218_010 |
0.1711705982 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 15 |
Hb_001900_060 |
0.1732106102 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 16 |
Hb_029977_010 |
0.1741280618 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_000540_040 |
0.176597999 |
- |
- |
PREDICTED: chitinase 2 [Amborella trichopoda] |
| 18 |
Hb_009535_030 |
0.1774260703 |
- |
- |
CP2 [Hevea brasiliensis] |
| 19 |
Hb_001663_120 |
0.1786714053 |
- |
- |
PREDICTED: formin-like protein 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001247_270 |
0.1788518568 |
- |
- |
PREDICTED: probable L-cysteine desulfhydrase, chloroplastic [Jatropha curcas] |