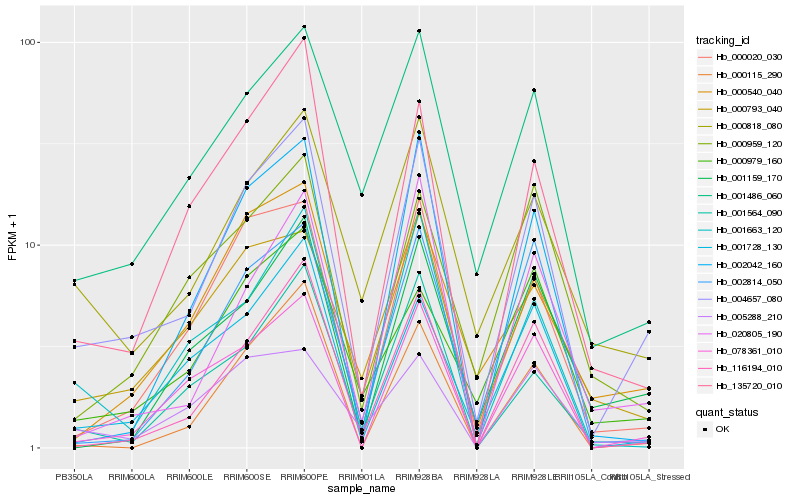
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004657_080 |
0.0 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 2 |
Hb_078361_010 |
0.1471259534 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 3 |
Hb_001486_060 |
0.1487011641 |
- |
- |
PREDICTED: uncharacterized protein LOC105632622 [Jatropha curcas] |
| 4 |
Hb_000020_030 |
0.1491141852 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_000818_080 |
0.1502102498 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 6 |
Hb_001728_130 |
0.1516951626 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_116194_010 |
0.153044597 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 8 |
Hb_135720_010 |
0.1548177344 |
- |
- |
hypothetical protein JCGZ_17857 [Jatropha curcas] |
| 9 |
Hb_001663_120 |
0.1635257233 |
- |
- |
PREDICTED: formin-like protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000793_040 |
0.165293155 |
- |
- |
PREDICTED: transcription factor LHW [Jatropha curcas] |
| 11 |
Hb_001159_170 |
0.1674745315 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 12 |
Hb_000540_040 |
0.16935328 |
- |
- |
PREDICTED: chitinase 2 [Amborella trichopoda] |
| 13 |
Hb_020805_190 |
0.1699215406 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_001564_090 |
0.172938065 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002042_160 |
0.1734821831 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 16 |
Hb_000115_290 |
0.1740856445 |
- |
- |
PREDICTED: uncharacterized protein LOC105124123 [Populus euphratica] |
| 17 |
Hb_002814_050 |
0.174387539 |
- |
- |
PREDICTED: sugar carrier protein A [Jatropha curcas] |
| 18 |
Hb_005288_210 |
0.1753692247 |
- |
- |
hypothetical protein VITISV_017217 [Vitis vinifera] |
| 19 |
Hb_000979_160 |
0.1755188673 |
- |
- |
hypothetical protein RCOM_0629030 [Ricinus communis] |
| 20 |
Hb_000959_120 |
0.1780667425 |
- |
- |
PREDICTED: protein BRASSINOSTEROID INSENSITIVE 1 [Jatropha curcas] |