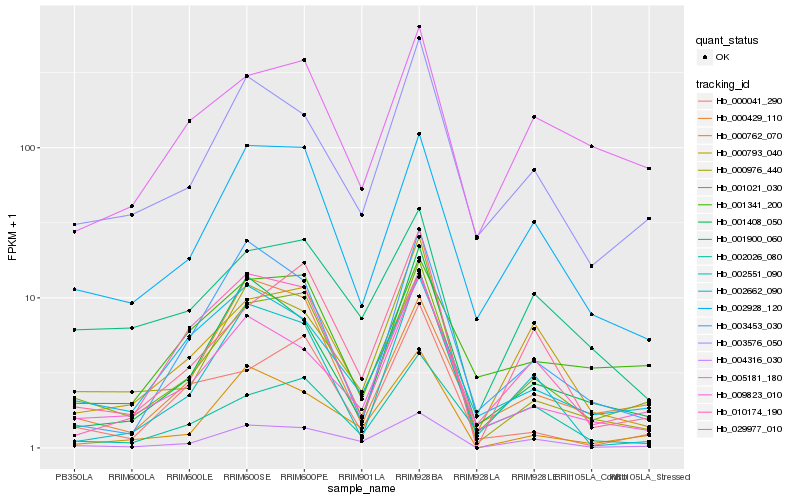
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001021_030 |
0.0 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 2 |
Hb_002928_120 |
0.119160834 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 3 |
Hb_003576_050 |
0.1351594447 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 4 |
Hb_004316_030 |
0.1416685003 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 5 |
Hb_029977_010 |
0.1463071898 |
- |
- |
catalytic, putative [Ricinus communis] |
| 6 |
Hb_009823_010 |
0.1523125477 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 7 |
Hb_000041_290 |
0.1536429252 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 8 |
Hb_001408_050 |
0.1553241567 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000976_440 |
0.1573487358 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_001341_200 |
0.1582744162 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 4 [Jatropha curcas] |
| 11 |
Hb_000793_040 |
0.1591277615 |
- |
- |
PREDICTED: transcription factor LHW [Jatropha curcas] |
| 12 |
Hb_005181_180 |
0.1602561119 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002026_080 |
0.1612999383 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 14 |
Hb_002662_090 |
0.1686029242 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 15 |
Hb_002551_090 |
0.1695981026 |
- |
- |
hypothetical protein JCGZ_17224 [Jatropha curcas] |
| 16 |
Hb_003453_030 |
0.1697163768 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 17 |
Hb_000762_070 |
0.1711799566 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 18 |
Hb_001900_060 |
0.1719661634 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 19 |
Hb_000429_110 |
0.1727819268 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2-like [Populus euphratica] |
| 20 |
Hb_010174_190 |
0.1747448319 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |