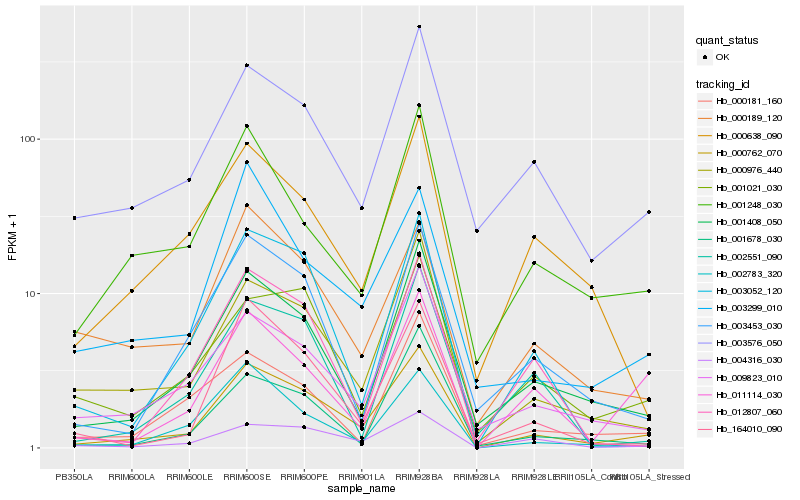
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000762_070 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 2 |
Hb_000976_440 |
0.1341326551 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_001408_050 |
0.1360839978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004316_030 |
0.1435721983 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 5 |
Hb_009823_010 |
0.1510154969 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 6 |
Hb_003576_050 |
0.1550570982 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 7 |
Hb_164010_090 |
0.1629645042 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 8 |
Hb_001248_030 |
0.1681390773 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_003453_030 |
0.1698358387 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 10 |
Hb_001021_030 |
0.1711799566 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 11 |
Hb_003052_120 |
0.1726351788 |
- |
- |
hypothetical protein POPTR_0008s10970g [Populus trichocarpa] |
| 12 |
Hb_011114_030 |
0.1765312549 |
- |
- |
PREDICTED: probable carboxylesterase 15 [Jatropha curcas] |
| 13 |
Hb_003299_010 |
0.1799642042 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 14 |
Hb_000181_160 |
0.1806827457 |
- |
- |
PREDICTED: uncharacterized protein LOC105637575 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001678_030 |
0.1807168569 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 16 |
Hb_002783_320 |
0.1812466179 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FC isoform X1 [Jatropha curcas] |
| 17 |
Hb_000189_120 |
0.182890269 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like isoform X5 [Jatropha curcas] |
| 18 |
Hb_000638_090 |
0.1837373689 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_012807_060 |
0.1845874486 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_002551_090 |
0.1847661898 |
- |
- |
hypothetical protein JCGZ_17224 [Jatropha curcas] |