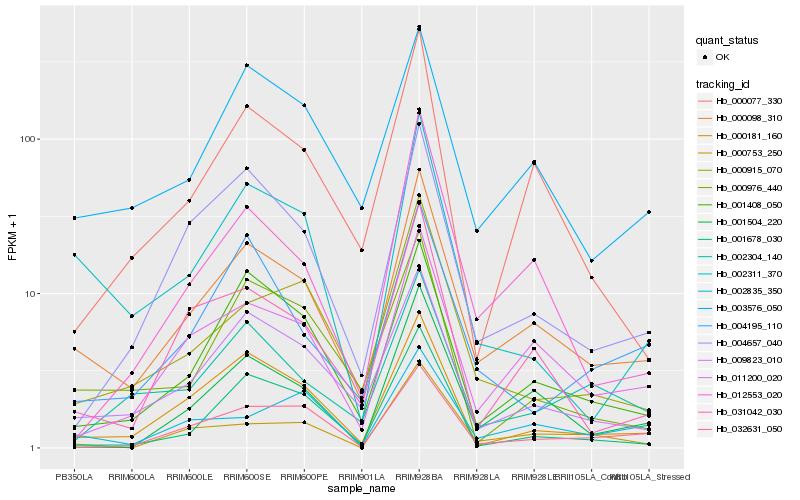
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000098_310 |
0.0 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 2 |
Hb_009823_010 |
0.1197832124 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 3 |
Hb_011200_020 |
0.1226424246 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 4 |
Hb_003576_050 |
0.1337166542 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 5 |
Hb_000181_160 |
0.1397788823 |
- |
- |
PREDICTED: uncharacterized protein LOC105637575 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001408_050 |
0.1477702652 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_031042_030 |
0.1500382916 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000077_330 |
0.1558482424 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2-like [Jatropha curcas] |
| 9 |
Hb_000915_070 |
0.1569892831 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_001504_220 |
0.1589753501 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_002311_370 |
0.159784566 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3 [Hevea brasiliensis] |
| 12 |
Hb_004657_040 |
0.1632919913 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 13 |
Hb_012553_020 |
0.1648249085 |
- |
- |
hypothetical protein JCGZ_09282 [Jatropha curcas] |
| 14 |
Hb_000753_250 |
0.16671993 |
- |
- |
PREDICTED: uncharacterized protein LOC105640661 [Jatropha curcas] |
| 15 |
Hb_032631_050 |
0.1688306534 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 16 |
Hb_002304_140 |
0.1727722864 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 17 |
Hb_000976_440 |
0.1738768532 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_001678_030 |
0.1743333792 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 19 |
Hb_004195_110 |
0.1768229056 |
- |
- |
hypothetical protein JCGZ_24010 [Jatropha curcas] |
| 20 |
Hb_002835_350 |
0.1783336747 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |