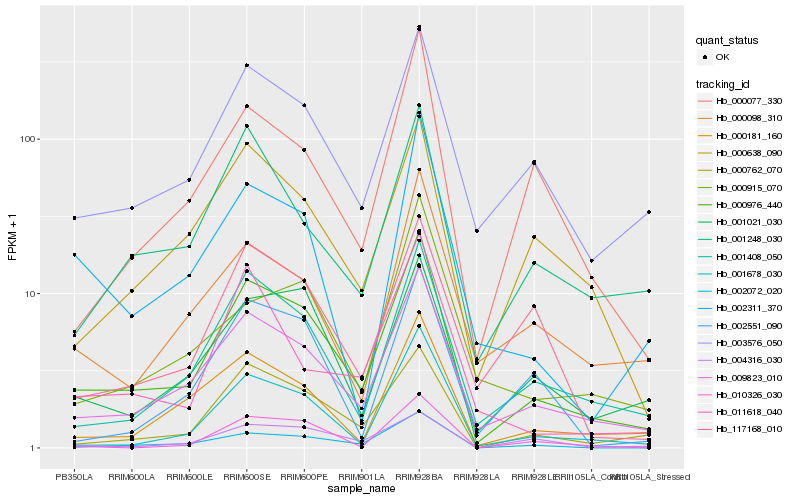
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000976_440 |
0.0 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_009823_010 |
0.0906234609 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 3 |
Hb_001408_050 |
0.1332317336 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000762_070 |
0.1341326551 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 5 |
Hb_002072_020 |
0.1386562189 |
- |
- |
hypothetical protein JCGZ_16057 [Jatropha curcas] |
| 6 |
Hb_003576_050 |
0.138768672 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 7 |
Hb_001678_030 |
0.1432402715 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 8 |
Hb_000181_160 |
0.1504908708 |
- |
- |
PREDICTED: uncharacterized protein LOC105637575 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001021_030 |
0.1573487358 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 10 |
Hb_000638_090 |
0.1618168523 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_000077_330 |
0.1660625009 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2-like [Jatropha curcas] |
| 12 |
Hb_002311_370 |
0.16866951 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3 [Hevea brasiliensis] |
| 13 |
Hb_001248_030 |
0.1693886644 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_004316_030 |
0.1714445401 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 15 |
Hb_000098_310 |
0.1738768532 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 16 |
Hb_000915_070 |
0.1820417658 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_010326_030 |
0.1836244812 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 4-like [Jatropha curcas] |
| 18 |
Hb_117168_010 |
0.1839620693 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 19 |
Hb_002551_090 |
0.1854541786 |
- |
- |
hypothetical protein JCGZ_17224 [Jatropha curcas] |
| 20 |
Hb_011618_040 |
0.1856673108 |
- |
- |
conserved hypothetical protein [Ricinus communis] |