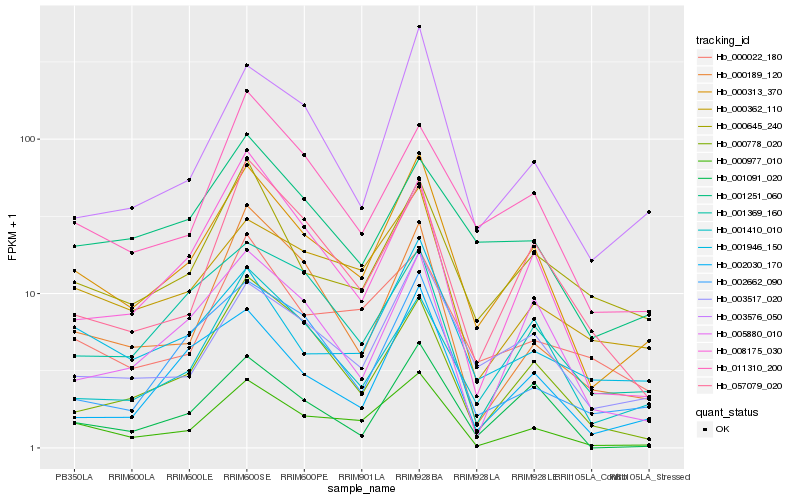
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000313_370 |
0.0 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_000977_010 |
0.1071454293 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 3 |
Hb_003517_020 |
0.1118315927 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 3-like [Jatropha curcas] |
| 4 |
Hb_003576_050 |
0.1322780147 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 5 |
Hb_002030_170 |
0.1382581926 |
- |
- |
PREDICTED: calcium-transporting ATPase 1-like [Jatropha curcas] |
| 6 |
Hb_001946_150 |
0.1413813058 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000362_110 |
0.1422216911 |
- |
- |
PREDICTED: uncharacterized protein LOC104612097 isoform X2 [Nelumbo nucifera] |
| 8 |
Hb_001091_020 |
0.1444742194 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 9 |
Hb_000645_240 |
0.1451214014 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 10 |
Hb_008175_030 |
0.1452719206 |
transcription factor |
TF Family: TAZ |
PREDICTED: BTB/POZ and TAZ domain-containing protein 3 [Jatropha curcas] |
| 11 |
Hb_011310_200 |
0.1472716362 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 12 |
Hb_005880_010 |
0.1502353866 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 13 |
Hb_002662_090 |
0.1538815506 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 14 |
Hb_001251_060 |
0.1568791525 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 15 |
Hb_000022_180 |
0.1576943783 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001369_160 |
0.1580519724 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 17 |
Hb_001410_010 |
0.1596324356 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP7-like [Jatropha curcas] |
| 18 |
Hb_000189_120 |
0.1600408365 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like isoform X5 [Jatropha curcas] |
| 19 |
Hb_000778_020 |
0.1611843606 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 20 |
Hb_057079_020 |
0.1614365396 |
- |
- |
conserved hypothetical protein [Ricinus communis] |