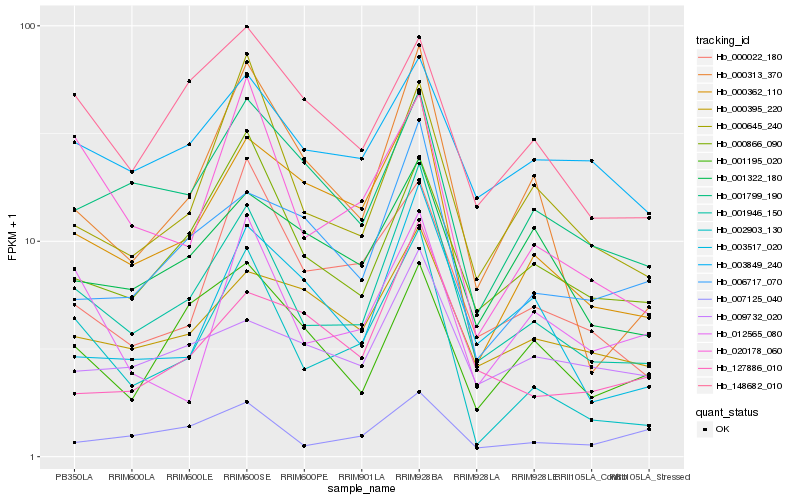
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001946_150 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002903_130 |
0.1312102429 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 3 |
Hb_000362_110 |
0.1377701089 |
- |
- |
PREDICTED: uncharacterized protein LOC104612097 isoform X2 [Nelumbo nucifera] |
| 4 |
Hb_000313_370 |
0.1413813058 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_000866_090 |
0.1478422455 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 6 |
Hb_000645_240 |
0.1484040536 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 7 |
Hb_001799_190 |
0.1560615011 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 8 |
Hb_006717_070 |
0.1570130492 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 9 |
Hb_003517_020 |
0.1588815884 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 3-like [Jatropha curcas] |
| 10 |
Hb_020178_060 |
0.161602609 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 3 [Jatropha curcas] |
| 11 |
Hb_000395_220 |
0.1634424964 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 12 |
Hb_009732_020 |
0.1654778157 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 13 |
Hb_000022_180 |
0.1665626618 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007125_040 |
0.1666479002 |
- |
- |
PREDICTED: cationic amino acid transporter 1-like [Jatropha curcas] |
| 15 |
Hb_003849_240 |
0.1671024495 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_001322_180 |
0.1682463511 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 17 |
Hb_148682_010 |
0.1713241477 |
- |
- |
PREDICTED: uncharacterized protein LOC105630720 [Jatropha curcas] |
| 18 |
Hb_127886_010 |
0.1737330616 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 19 |
Hb_001195_020 |
0.1739032162 |
- |
- |
PREDICTED: ornithine carbamoyltransferase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_012565_080 |
0.1764606479 |
- |
- |
serine/threonine kinase, putative [Ricinus communis] |