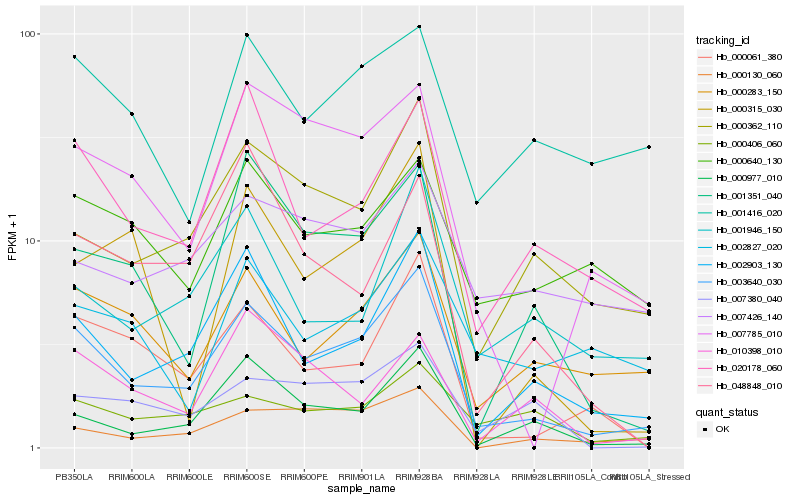
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003640_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002903_130 |
0.1329604705 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 3 |
Hb_000362_110 |
0.157839304 |
- |
- |
PREDICTED: uncharacterized protein LOC104612097 isoform X2 [Nelumbo nucifera] |
| 4 |
Hb_020178_060 |
0.1645472966 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 3 [Jatropha curcas] |
| 5 |
Hb_000283_150 |
0.1649354996 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 6 |
Hb_000977_010 |
0.1658346404 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 7 |
Hb_000061_380 |
0.1697348828 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_007785_010 |
0.1773741083 |
- |
- |
glutathione peroxidase, putative [Ricinus communis] |
| 9 |
Hb_001351_040 |
0.1775392303 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 2, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_000130_060 |
0.1838460458 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 11 |
Hb_001946_150 |
0.1914444944 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000406_060 |
0.1933936147 |
- |
- |
hypothetical protein CICLE_v10000115mg [Citrus clementina] |
| 13 |
Hb_007380_040 |
0.1968738457 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 14 |
Hb_000640_130 |
0.2029436582 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_048848_010 |
0.2055048576 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 16 |
Hb_001416_020 |
0.2077553243 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 17 |
Hb_007426_140 |
0.2080454455 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 18 |
Hb_010398_010 |
0.2088895217 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: mediator of RNA polymerase II transcription subunit 15a-like [Malus domestica] |
| 19 |
Hb_002827_020 |
0.2092286781 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 20 |
Hb_000315_030 |
0.2119989868 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |