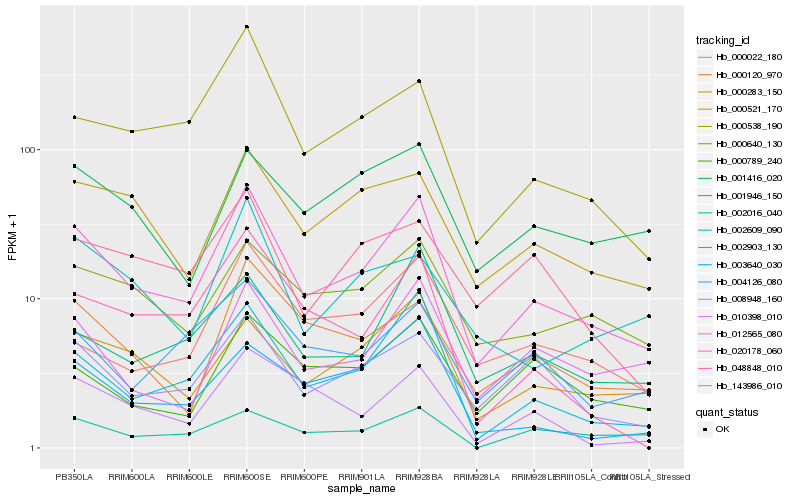
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020178_060 |
0.0 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 3 [Jatropha curcas] |
| 2 |
Hb_002903_130 |
0.1212297571 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 3 |
Hb_000283_150 |
0.1414223145 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 4 |
Hb_008948_160 |
0.1424569005 |
- |
- |
PREDICTED: uncharacterized protein LOC104214688 isoform X2 [Nicotiana sylvestris] |
| 5 |
Hb_012565_080 |
0.1484564562 |
- |
- |
serine/threonine kinase, putative [Ricinus communis] |
| 6 |
Hb_000538_190 |
0.1520912492 |
- |
- |
PREDICTED: protein DJ-1 homolog D [Jatropha curcas] |
| 7 |
Hb_000521_170 |
0.1539259366 |
- |
- |
PREDICTED: callose synthase 1 [Jatropha curcas] |
| 8 |
Hb_000640_130 |
0.1548296499 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_000120_970 |
0.1560541257 |
- |
- |
PREDICTED: probable acyl-activating enzyme 2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001946_150 |
0.161602609 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004126_080 |
0.1624723122 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |
| 12 |
Hb_003640_030 |
0.1645472966 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000022_180 |
0.1688749087 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000789_240 |
0.1694999211 |
- |
- |
gamma glutamyl transpeptidases, putative [Ricinus communis] |
| 15 |
Hb_001416_020 |
0.1736477301 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 16 |
Hb_048848_010 |
0.1744217681 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 17 |
Hb_143986_010 |
0.174855713 |
- |
- |
PREDICTED: beta-galactosidase [Jatropha curcas] |
| 18 |
Hb_002016_040 |
0.1749493072 |
rubber biosynthesis |
Gene Name: SRPP8 |
PREDICTED: stress-related protein [Jatropha curcas] |
| 19 |
Hb_010398_010 |
0.1755347747 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: mediator of RNA polymerase II transcription subunit 15a-like [Malus domestica] |
| 20 |
Hb_002609_090 |
0.1758081149 |
- |
- |
PREDICTED: phosphomethylpyrimidine synthase, chloroplastic isoform X2 [Jatropha curcas] |