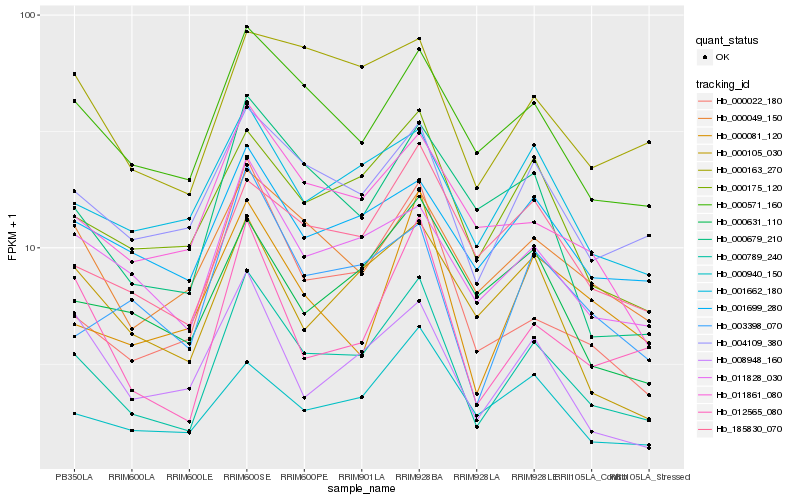
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000789_240 |
0.0 |
- |
- |
gamma glutamyl transpeptidases, putative [Ricinus communis] |
| 2 |
Hb_000022_180 |
0.1243550086 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000175_120 |
0.1251542111 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 4 |
Hb_012565_080 |
0.1254792233 |
- |
- |
serine/threonine kinase, putative [Ricinus communis] |
| 5 |
Hb_004109_380 |
0.1260848889 |
- |
- |
PREDICTED: callose synthase 9 isoform X4 [Pyrus x bretschneideri] |
| 6 |
Hb_000571_160 |
0.1298386447 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 7 |
Hb_011828_030 |
0.1307439438 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 8 |
Hb_000679_210 |
0.136429084 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 9 |
Hb_185830_070 |
0.1402226327 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_011861_080 |
0.1409344915 |
- |
- |
hypothetical protein JCGZ_06088 [Jatropha curcas] |
| 11 |
Hb_003398_070 |
0.1420738971 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 12 |
Hb_000049_150 |
0.1422170272 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_000940_150 |
0.1439699475 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 14 |
Hb_001662_180 |
0.1454739181 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 15 |
Hb_000081_120 |
0.1496838106 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 16 |
Hb_000105_030 |
0.1503237294 |
- |
- |
fkbp-rapamycin associated protein, putative [Ricinus communis] |
| 17 |
Hb_000163_270 |
0.1512659194 |
- |
- |
PREDICTED: uncharacterized protein LOC105642512 [Jatropha curcas] |
| 18 |
Hb_008948_160 |
0.1525356377 |
- |
- |
PREDICTED: uncharacterized protein LOC104214688 isoform X2 [Nicotiana sylvestris] |
| 19 |
Hb_001699_280 |
0.1530379667 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 20 |
Hb_000631_110 |
0.153279045 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 3 [Jatropha curcas] |