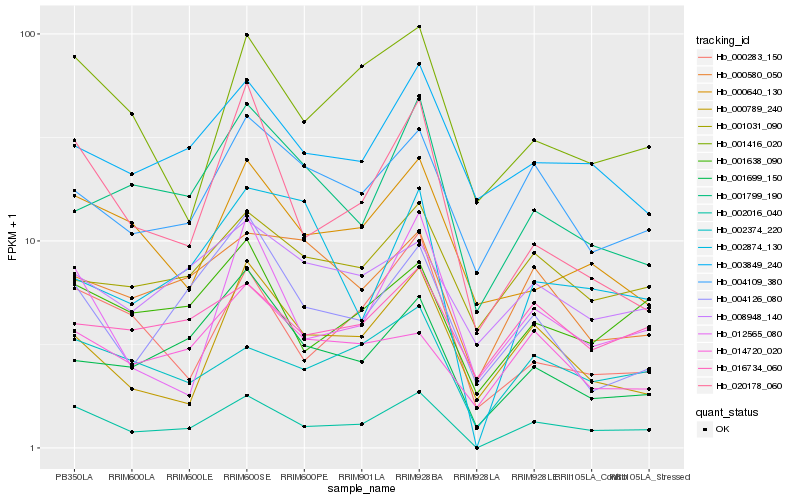
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002016_040 |
0.0 |
rubber biosynthesis |
Gene Name: SRPP8 |
PREDICTED: stress-related protein [Jatropha curcas] |
| 2 |
Hb_002374_220 |
0.1561829531 |
- |
- |
hypothetical protein CICLE_v10001597mg [Citrus clementina] |
| 3 |
Hb_001638_090 |
0.1572998642 |
- |
- |
GRF1-INTERACTING FACTOR 2 family protein [Populus trichocarpa] |
| 4 |
Hb_012565_080 |
0.1711913562 |
- |
- |
serine/threonine kinase, putative [Ricinus communis] |
| 5 |
Hb_004109_380 |
0.1730096298 |
- |
- |
PREDICTED: callose synthase 9 isoform X4 [Pyrus x bretschneideri] |
| 6 |
Hb_000283_150 |
0.1732505331 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 7 |
Hb_020178_060 |
0.1749493072 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 3 [Jatropha curcas] |
| 8 |
Hb_001416_020 |
0.1750630695 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 9 |
Hb_001699_150 |
0.1770728986 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 10 |
Hb_000640_130 |
0.1808937365 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_001031_090 |
0.1821437825 |
- |
- |
PREDICTED: translational activator GCN1 [Jatropha curcas] |
| 12 |
Hb_003849_240 |
0.1827666782 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_016734_060 |
0.1828887851 |
- |
- |
PREDICTED: nucleolar protein 6 [Jatropha curcas] |
| 14 |
Hb_004126_080 |
0.1831887597 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |
| 15 |
Hb_000789_240 |
0.1835958172 |
- |
- |
gamma glutamyl transpeptidases, putative [Ricinus communis] |
| 16 |
Hb_000580_050 |
0.1855729753 |
- |
- |
- |
| 17 |
Hb_001799_190 |
0.1870614008 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 18 |
Hb_008948_140 |
0.1871451644 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |
| 19 |
Hb_014720_020 |
0.1878468829 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM [Jatropha curcas] |
| 20 |
Hb_002874_130 |
0.1884650158 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |