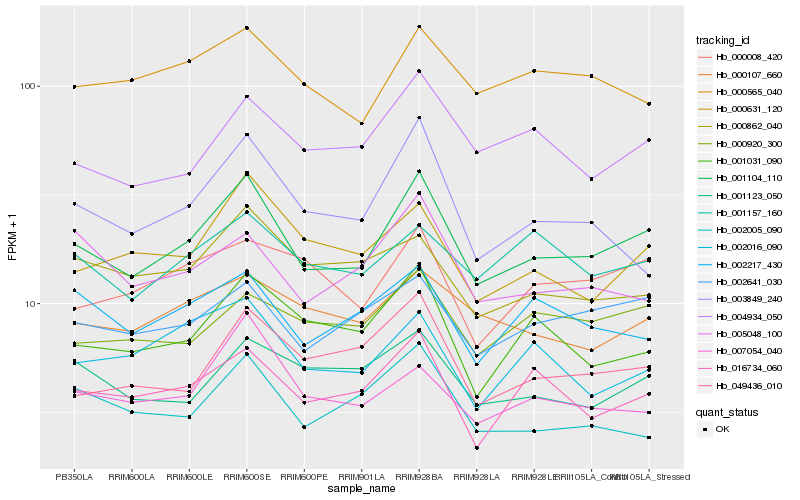
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001104_110 |
0.0 |
- |
- |
PREDICTED: splicing factor U2af small subunit B-like [Populus euphratica] |
| 2 |
Hb_002641_030 |
0.0941048991 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X1 [Jatropha curcas] |
| 3 |
Hb_004934_050 |
0.0971265426 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 4 |
Hb_016734_060 |
0.0985075447 |
- |
- |
PREDICTED: nucleolar protein 6 [Jatropha curcas] |
| 5 |
Hb_000631_120 |
0.1018478246 |
- |
- |
hypothetical protein CISIN_1g0431902mg, partial [Citrus sinensis] |
| 6 |
Hb_049436_010 |
0.1033848471 |
- |
- |
PREDICTED: UPF0202 protein At1g10490-like [Glycine max] |
| 7 |
Hb_001031_090 |
0.1041472134 |
- |
- |
PREDICTED: translational activator GCN1 [Jatropha curcas] |
| 8 |
Hb_000920_300 |
0.1047041416 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 9 |
Hb_000565_040 |
0.1053569696 |
- |
- |
RecName: Full=Superoxide dismutase [Mn], mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 10 |
Hb_001123_050 |
0.1072147474 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000008_420 |
0.1076330151 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003849_240 |
0.1076478329 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_002217_430 |
0.1080904788 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X4 [Jatropha curcas] |
| 14 |
Hb_000107_660 |
0.1082729127 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 15 |
Hb_007054_040 |
0.1099449333 |
- |
- |
PREDICTED: protein RST1 [Jatropha curcas] |
| 16 |
Hb_001157_160 |
0.1100450302 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002005_090 |
0.1114401554 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002016_090 |
0.1127302604 |
- |
- |
PREDICTED: DNA mismatch repair protein PMS1 [Jatropha curcas] |
| 19 |
Hb_005048_100 |
0.1127655435 |
- |
- |
secretory carrier membrane protein, putative [Ricinus communis] |
| 20 |
Hb_000862_040 |
0.112835169 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |