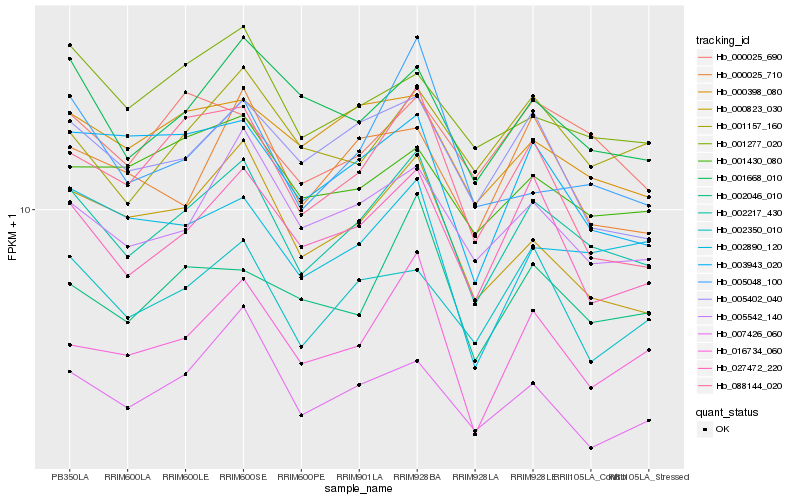
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002217_430 |
0.0 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X4 [Jatropha curcas] |
| 2 |
Hb_027472_220 |
0.0676328102 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_005542_140 |
0.0725813233 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 4 |
Hb_001668_010 |
0.0769318502 |
- |
- |
PREDICTED: RNA-binding protein with multiple splicing [Jatropha curcas] |
| 5 |
Hb_016734_060 |
0.0773814439 |
- |
- |
PREDICTED: nucleolar protein 6 [Jatropha curcas] |
| 6 |
Hb_000398_080 |
0.0794847136 |
- |
- |
tip120, putative [Ricinus communis] |
| 7 |
Hb_002350_010 |
0.0806599527 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc1 [Jatropha curcas] |
| 8 |
Hb_088144_020 |
0.0809522956 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_005402_040 |
0.0820011803 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 10 |
Hb_001277_020 |
0.082291441 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 11 |
Hb_000823_030 |
0.0826352577 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 12 |
Hb_002046_010 |
0.0826745254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000025_690 |
0.0846997392 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 14 |
Hb_001430_080 |
0.0858072933 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 15 |
Hb_005048_100 |
0.0858483438 |
- |
- |
secretory carrier membrane protein, putative [Ricinus communis] |
| 16 |
Hb_001157_160 |
0.0863591105 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003943_020 |
0.08663702 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002890_120 |
0.086907024 |
- |
- |
Nucleic acid binding,RNA binding isoform 1 [Theobroma cacao] |
| 19 |
Hb_007426_060 |
0.0872683657 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_000025_710 |
0.0881253653 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |