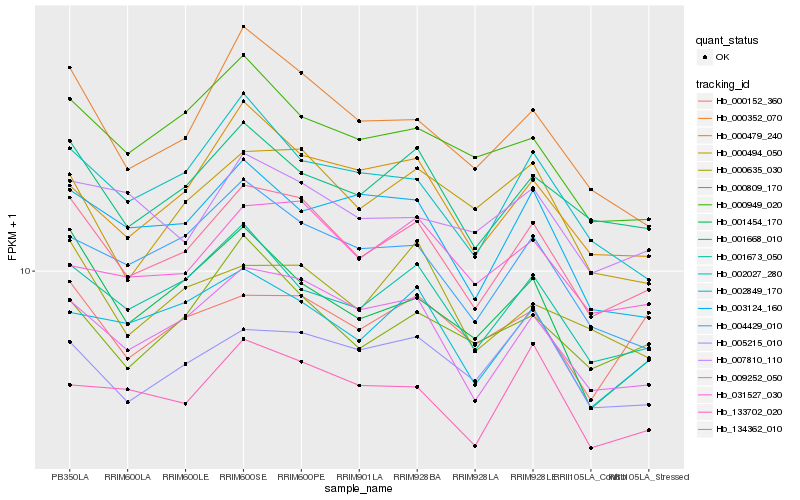
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_134362_010 |
0.0 |
- |
- |
PREDICTED: FAS-associated factor 2 [Jatropha curcas] |
| 2 |
Hb_009252_050 |
0.0532521189 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_001668_010 |
0.0597116329 |
- |
- |
PREDICTED: RNA-binding protein with multiple splicing [Jatropha curcas] |
| 4 |
Hb_001673_050 |
0.0649816351 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_002849_170 |
0.0739843302 |
- |
- |
PREDICTED: uncharacterized protein LOC105642955 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000352_070 |
0.0740291205 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 7 |
Hb_004429_010 |
0.0766234239 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 8 |
Hb_002027_280 |
0.0791642073 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 9 |
Hb_000479_240 |
0.0796843349 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001454_170 |
0.0806235627 |
- |
- |
PREDICTED: uncharacterized protein LOC105643431 [Jatropha curcas] |
| 11 |
Hb_133702_020 |
0.0806337052 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_031527_030 |
0.0809666101 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 13 |
Hb_000635_030 |
0.0809886084 |
- |
- |
mannosyl-oligosaccharide glucosidase, putative [Ricinus communis] |
| 14 |
Hb_000809_170 |
0.0821361111 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein 1 homolog isoform X1 [Jatropha curcas] |
| 15 |
Hb_000494_050 |
0.0824180787 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 16 |
Hb_005215_010 |
0.0831625695 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000949_020 |
0.0833070377 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 18 |
Hb_003124_160 |
0.0839034161 |
- |
- |
dynamin, putative [Ricinus communis] |
| 19 |
Hb_000152_360 |
0.0840945142 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP53 [Jatropha curcas] |
| 20 |
Hb_007810_110 |
0.0852326623 |
- |
- |
PREDICTED: DNA/RNA-binding protein KIN17 [Jatropha curcas] |