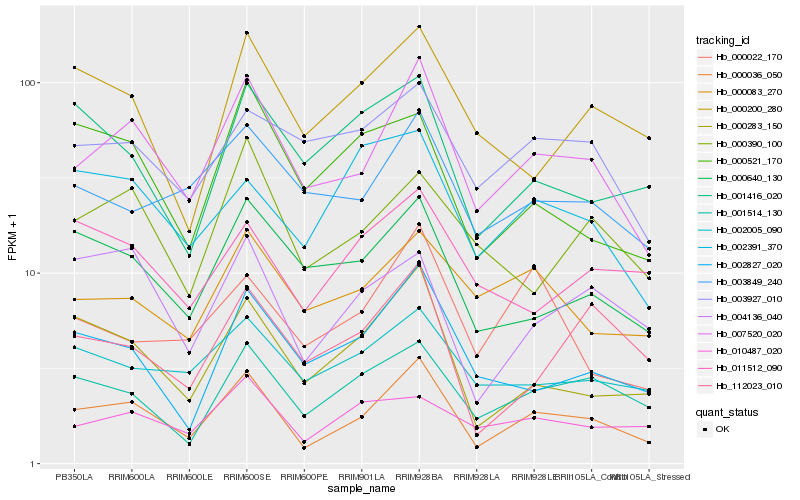
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000036_050 |
0.0 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 6-like [Camelina sativa] |
| 2 |
Hb_007520_020 |
0.0963486452 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
1-deoxy-D-xylulose-5-phosphate reductoisomerase [Hevea brasiliensis] |
| 3 |
Hb_000283_150 |
0.1410156354 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 4 |
Hb_001514_130 |
0.1499964615 |
- |
- |
hypothetical protein JCGZ_16073 [Jatropha curcas] |
| 5 |
Hb_004136_040 |
0.1579834433 |
- |
- |
putative protein kinase [Arabidopsis thaliana] |
| 6 |
Hb_002827_020 |
0.1635603438 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 7 |
Hb_000640_130 |
0.1653495922 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_002005_090 |
0.1717680851 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_112023_010 |
0.1745470114 |
- |
- |
hypothetical protein EUGRSUZ_C03826 [Eucalyptus grandis] |
| 10 |
Hb_003849_240 |
0.1759047271 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000390_100 |
0.1787367138 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 12 |
Hb_000200_280 |
0.1797033592 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 13 |
Hb_000083_270 |
0.1797156514 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 14 |
Hb_010487_020 |
0.1822819188 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 15 |
Hb_001416_020 |
0.1824451399 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 16 |
Hb_000521_170 |
0.1835598159 |
- |
- |
PREDICTED: callose synthase 1 [Jatropha curcas] |
| 17 |
Hb_002391_370 |
0.1842310745 |
- |
- |
PREDICTED: aconitate hydratase 1 [Jatropha curcas] |
| 18 |
Hb_003927_010 |
0.1859839554 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_011512_090 |
0.1862942084 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 20 |
Hb_000022_170 |
0.1864584988 |
- |
- |
PREDICTED: uncharacterized protein LOC105628473 [Jatropha curcas] |