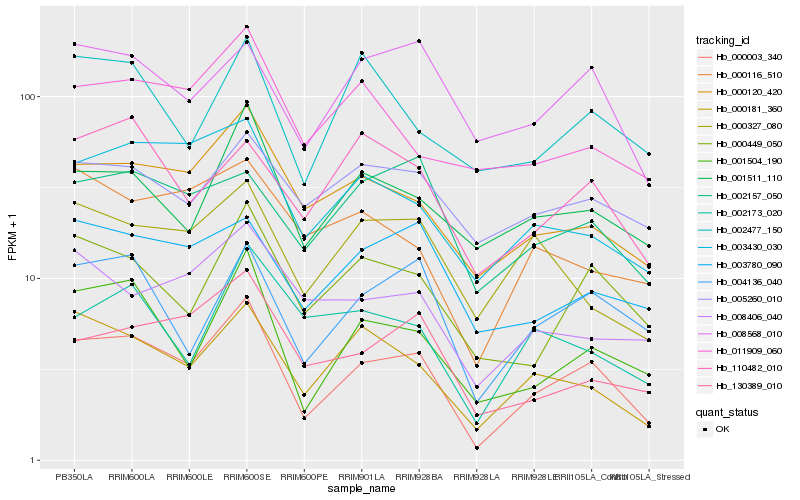
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_340 |
0.0 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |
| 2 |
Hb_001504_190 |
0.1312786812 |
- |
- |
double-stranded RNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_000120_420 |
0.1406047941 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |
| 4 |
Hb_000181_360 |
0.143380521 |
- |
- |
PREDICTED: uncharacterized protein LOC105644273 [Jatropha curcas] |
| 5 |
Hb_004136_040 |
0.143541437 |
- |
- |
putative protein kinase [Arabidopsis thaliana] |
| 6 |
Hb_000449_050 |
0.1439446122 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 7 |
Hb_001511_110 |
0.1531888033 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003430_030 |
0.1531890479 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 9 |
Hb_130389_010 |
0.1607708829 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 10 |
Hb_003780_090 |
0.1637672276 |
- |
- |
PREDICTED: transcription factor bHLH128 [Jatropha curcas] |
| 11 |
Hb_008568_010 |
0.164991086 |
- |
- |
Phosphoenolpyruvate carboxykinase [ATP], putative [Ricinus communis] |
| 12 |
Hb_000327_080 |
0.1688211945 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 13 |
Hb_002157_050 |
0.1690950685 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 14 |
Hb_110482_010 |
0.1718119905 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 15 |
Hb_008406_040 |
0.1733247512 |
- |
- |
- |
| 16 |
Hb_000116_510 |
0.1748583972 |
- |
- |
PREDICTED: RNA-binding protein 24-A-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_011909_060 |
0.1751235904 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 18 |
Hb_005260_010 |
0.1751751123 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002173_020 |
0.1762161281 |
- |
- |
PREDICTED: uncharacterized protein LOC105801078 [Gossypium raimondii] |
| 20 |
Hb_002477_150 |
0.1776793669 |
transcription factor |
TF Family: Alfin-like |
DNA binding protein, putative [Ricinus communis] |