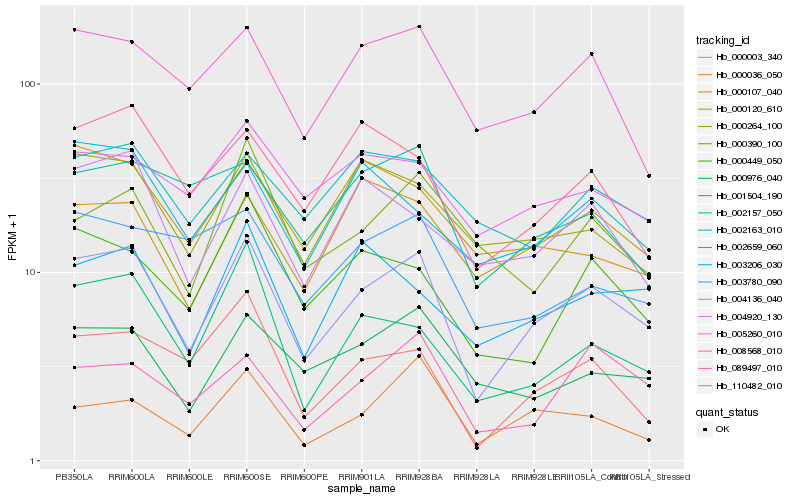
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004136_040 |
0.0 |
- |
- |
putative protein kinase [Arabidopsis thaliana] |
| 2 |
Hb_008568_010 |
0.1351135101 |
- |
- |
Phosphoenolpyruvate carboxykinase [ATP], putative [Ricinus communis] |
| 3 |
Hb_000976_040 |
0.139603106 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 4 |
Hb_000003_340 |
0.143541437 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |
| 5 |
Hb_000120_610 |
0.1454207369 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 6 |
Hb_000264_100 |
0.1468368413 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001504_190 |
0.14708596 |
- |
- |
double-stranded RNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_004920_130 |
0.1485365136 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_089497_010 |
0.1485412496 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |
| 10 |
Hb_000449_050 |
0.1489355801 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 11 |
Hb_110482_010 |
0.1496760394 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 12 |
Hb_005260_010 |
0.1497464363 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003206_030 |
0.1506385529 |
- |
- |
PREDICTED: uncharacterized protein LOC105632652 [Jatropha curcas] |
| 14 |
Hb_002163_010 |
0.1525391092 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410 [Jatropha curcas] |
| 15 |
Hb_002157_050 |
0.1533359426 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 16 |
Hb_002659_060 |
0.1552331535 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3B [Jatropha curcas] |
| 17 |
Hb_003780_090 |
0.1557336741 |
- |
- |
PREDICTED: transcription factor bHLH128 [Jatropha curcas] |
| 18 |
Hb_000107_040 |
0.1572433027 |
- |
- |
nuclear pore complex protein nup153, putative [Ricinus communis] |
| 19 |
Hb_000390_100 |
0.1579758283 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 20 |
Hb_000036_050 |
0.1579834433 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 6-like [Camelina sativa] |