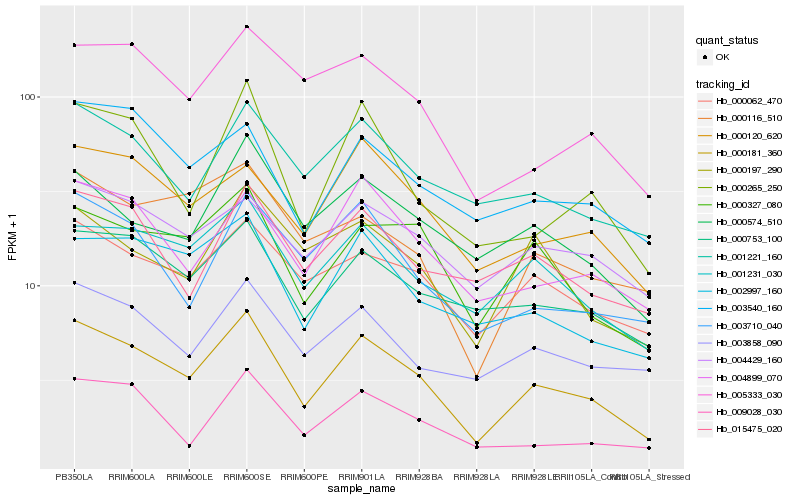
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000181_360 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644273 [Jatropha curcas] |
| 2 |
Hb_001231_030 |
0.1090151352 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 3 |
Hb_000327_080 |
0.1112223745 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 4 |
Hb_004899_070 |
0.1121201036 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 5 |
Hb_000265_250 |
0.1142863031 |
- |
- |
auxilin, putative [Ricinus communis] |
| 6 |
Hb_000062_470 |
0.1146226525 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 7 |
Hb_000753_100 |
0.1155864505 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000120_620 |
0.1159135265 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1a isoform X1 [Jatropha curcas] |
| 9 |
Hb_002997_160 |
0.1169767444 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001221_160 |
0.1186482462 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 11 |
Hb_003858_090 |
0.1209650973 |
- |
- |
Filamin-A-interacting protein, putative [Ricinus communis] |
| 12 |
Hb_003710_040 |
0.1221653575 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 13 |
Hb_009028_030 |
0.1235302519 |
desease resistance |
Gene Name: NB-ARC |
NBS type disease resistance protein, putative [Theobroma cacao] |
| 14 |
Hb_003540_160 |
0.1236786736 |
- |
- |
hypothetical protein POPTR_0011s12160g [Populus trichocarpa] |
| 15 |
Hb_004429_160 |
0.1250268635 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005333_030 |
0.1251357403 |
- |
- |
hypothetical protein JCGZ_15866 [Jatropha curcas] |
| 17 |
Hb_015475_020 |
0.1252973177 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 18 |
Hb_000197_290 |
0.125605428 |
- |
- |
PREDICTED: A-agglutinin anchorage subunit [Jatropha curcas] |
| 19 |
Hb_000574_510 |
0.1259217921 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 20 |
Hb_000116_510 |
0.1271534295 |
- |
- |
PREDICTED: RNA-binding protein 24-A-like isoform X2 [Jatropha curcas] |