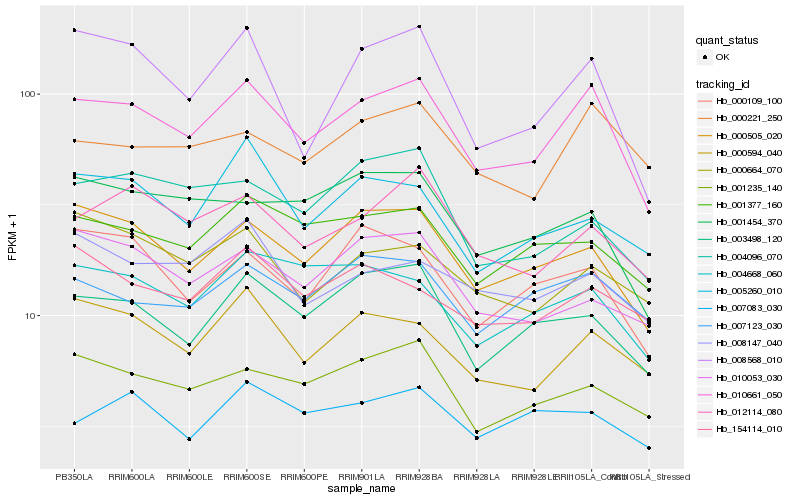
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010661_050 |
0.0 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP]-like [Fragaria vesca subsp. vesca] |
| 2 |
Hb_000505_020 |
0.0758305458 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 3 |
Hb_003498_120 |
0.0848695087 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 4 |
Hb_000594_040 |
0.0895655835 |
- |
- |
hypothetical protein L484_013922 [Morus notabilis] |
| 5 |
Hb_008568_010 |
0.0896849472 |
- |
- |
Phosphoenolpyruvate carboxykinase [ATP], putative [Ricinus communis] |
| 6 |
Hb_012114_080 |
0.0901281406 |
- |
- |
gamma-glutamylcysteine synthetase [Hevea brasiliensis] |
| 7 |
Hb_001235_140 |
0.0912064003 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.6 [Jatropha curcas] |
| 8 |
Hb_001377_160 |
0.0928864513 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 9 |
Hb_000221_250 |
0.0938296548 |
- |
- |
hypothetical protein JCGZ_15814 [Jatropha curcas] |
| 10 |
Hb_007123_030 |
0.0962832774 |
- |
- |
PREDICTED: uncharacterized protein LOC105634056 isoform X3 [Jatropha curcas] |
| 11 |
Hb_000109_100 |
0.0969920727 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 [Jatropha curcas] |
| 12 |
Hb_004668_060 |
0.0987758781 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001454_370 |
0.0992635522 |
- |
- |
PREDICTED: glyoxysomal fatty acid beta-oxidation multifunctional protein MFP-a [Jatropha curcas] |
| 14 |
Hb_005260_010 |
0.1045457425 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004096_070 |
0.1045630185 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_000664_070 |
0.1057125334 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 17 |
Hb_007083_030 |
0.1061871914 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 18 |
Hb_008147_040 |
0.1062039437 |
- |
- |
PREDICTED: protein FLX-like 2 [Jatropha curcas] |
| 19 |
Hb_010053_030 |
0.1073636293 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 20 |
Hb_154114_010 |
0.1080239595 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |