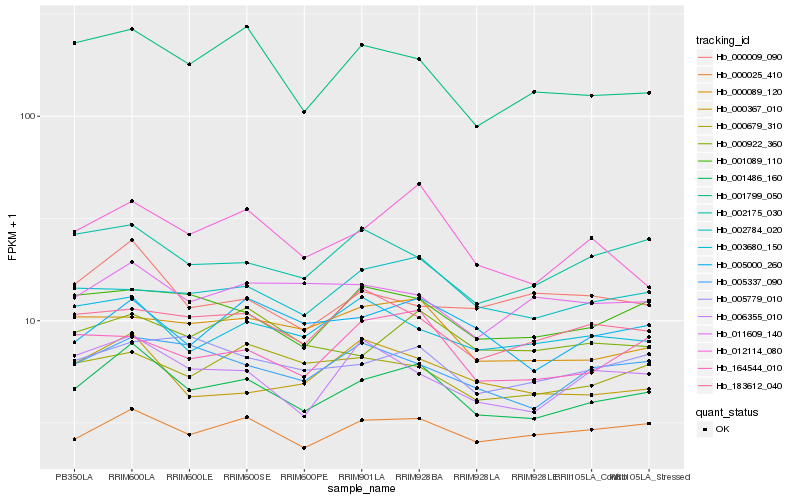
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001486_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 2 |
Hb_000025_410 |
0.0777182726 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g01110-like [Populus euphratica] |
| 3 |
Hb_005337_090 |
0.0854214919 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like [Jatropha curcas] |
| 4 |
Hb_001089_110 |
0.0866282835 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_012114_080 |
0.0889835758 |
- |
- |
gamma-glutamylcysteine synthetase [Hevea brasiliensis] |
| 6 |
Hb_005779_010 |
0.0891572119 |
- |
- |
catalytic, putative [Ricinus communis] |
| 7 |
Hb_005000_260 |
0.0901838158 |
- |
- |
recA family protein [Populus trichocarpa] |
| 8 |
Hb_006355_010 |
0.0905471283 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 9 |
Hb_000922_360 |
0.0915017227 |
- |
- |
- |
| 10 |
Hb_001799_050 |
0.0928647418 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 11 |
Hb_000089_120 |
0.0932088041 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 12 |
Hb_164544_010 |
0.0933070146 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.5 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002784_020 |
0.0959850032 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 3 [Jatropha curcas] |
| 14 |
Hb_002175_030 |
0.0961847668 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X2 [Jatropha curcas] |
| 15 |
Hb_000679_310 |
0.0970133993 |
- |
- |
PREDICTED: putative UDP-sugar transporter DDB_G0278631 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000367_010 |
0.0976112878 |
- |
- |
PREDICTED: RNA polymerase-associated protein LEO1 [Jatropha curcas] |
| 17 |
Hb_011609_140 |
0.0985188798 |
- |
- |
hypothetical protein JCGZ_12974 [Jatropha curcas] |
| 18 |
Hb_003680_150 |
0.0986709422 |
- |
- |
srpk, putative [Ricinus communis] |
| 19 |
Hb_000009_090 |
0.0986756409 |
transcription factor |
TF Family: bZIP |
G-box-binding factor, putative [Ricinus communis] |
| 20 |
Hb_183612_040 |
0.0990522819 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |