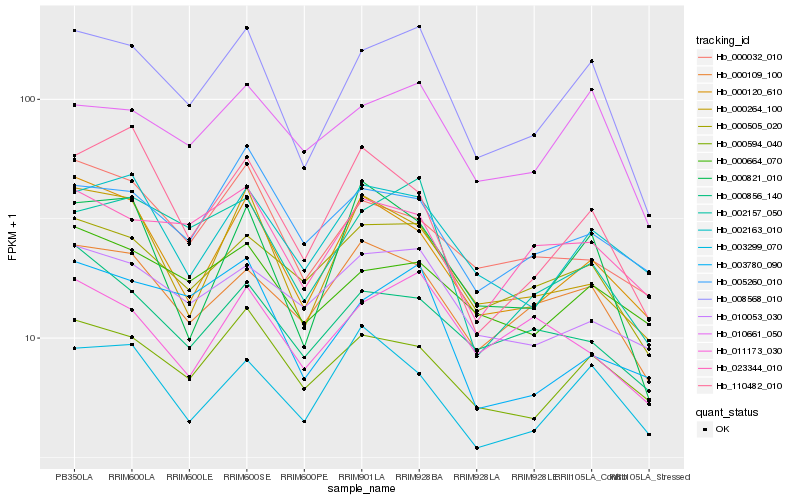
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008568_010 |
0.0 |
- |
- |
Phosphoenolpyruvate carboxykinase [ATP], putative [Ricinus communis] |
| 2 |
Hb_010661_050 |
0.0896849472 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP]-like [Fragaria vesca subsp. vesca] |
| 3 |
Hb_002157_050 |
0.090512573 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 4 |
Hb_000505_020 |
0.0945233011 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 5 |
Hb_000109_100 |
0.0996843291 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 [Jatropha curcas] |
| 6 |
Hb_000120_610 |
0.1039470411 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 7 |
Hb_000821_010 |
0.1061013671 |
- |
- |
PREDICTED: auxin response factor 19-like [Jatropha curcas] |
| 8 |
Hb_000594_040 |
0.1075254291 |
- |
- |
hypothetical protein L484_013922 [Morus notabilis] |
| 9 |
Hb_023344_010 |
0.1091555366 |
- |
- |
PREDICTED: pre-mRNA-processing factor 39 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000264_100 |
0.110095255 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002163_010 |
0.1108572593 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410 [Jatropha curcas] |
| 12 |
Hb_003780_090 |
0.1136576432 |
- |
- |
PREDICTED: transcription factor bHLH128 [Jatropha curcas] |
| 13 |
Hb_000664_070 |
0.1153549759 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 14 |
Hb_003299_070 |
0.1154636291 |
- |
- |
AMP deaminase [Theobroma cacao] |
| 15 |
Hb_005260_010 |
0.1163015578 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 16 |
Hb_010053_030 |
0.1181837184 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 17 |
Hb_000032_010 |
0.1186250819 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 18 |
Hb_000856_140 |
0.1199000442 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 19 |
Hb_110482_010 |
0.1204329094 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 20 |
Hb_011173_030 |
0.1208555053 |
- |
- |
pumilio, putative [Ricinus communis] |