| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
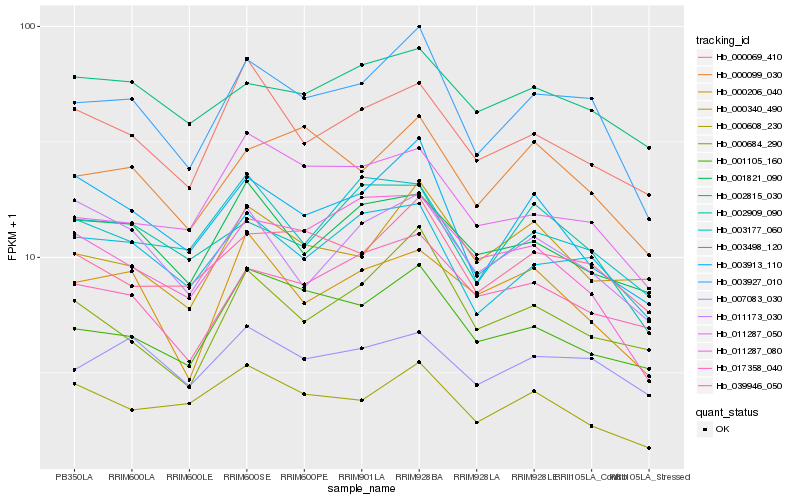
Hb_003927_010 |
0.0 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_003498_120 |
0.0821418779 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 3 |
Hb_039946_050 |
0.0907847916 |
- |
- |
catalytic, putative [Ricinus communis] |
| 4 |
Hb_000099_030 |
0.0962909066 |
- |
- |
6-phosphogluconate dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_003913_110 |
0.096807578 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 6 |
Hb_001105_160 |
0.1015424358 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 7 |
Hb_007083_030 |
0.1019244625 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 8 |
Hb_002909_090 |
0.1034936788 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase DPE2 [Jatropha curcas] |
| 9 |
Hb_017358_040 |
0.103562658 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 10 |
Hb_000069_410 |
0.1050435423 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 11 |
Hb_000684_290 |
0.1069688079 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 12 |
Hb_011287_050 |
0.1073122895 |
- |
- |
PREDICTED: uncharacterized protein LOC105644786 [Jatropha curcas] |
| 13 |
Hb_003177_060 |
0.1089662981 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001821_090 |
0.1089915551 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 15 |
Hb_002815_030 |
0.1098025702 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 16 |
Hb_000608_230 |
0.1101525803 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000340_490 |
0.1112166775 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 18 |
Hb_000206_040 |
0.111569477 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 19 |
Hb_011173_030 |
0.1129284638 |
- |
- |
pumilio, putative [Ricinus communis] |
| 20 |
Hb_011287_080 |
0.1130274284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |