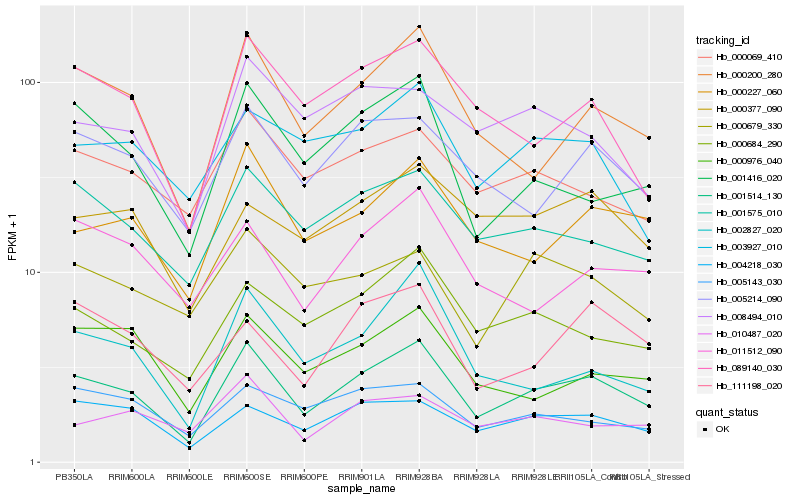
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001514_130 |
0.0 |
- |
- |
hypothetical protein JCGZ_16073 [Jatropha curcas] |
| 2 |
Hb_000200_280 |
0.1077155953 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 3 |
Hb_000227_060 |
0.1169735085 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 4 |
Hb_001575_010 |
0.1207702503 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_005214_090 |
0.1232534764 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 6 |
Hb_004218_030 |
0.1249464115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000684_290 |
0.1251360735 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 8 |
Hb_089140_030 |
0.1294099801 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 9 |
Hb_001416_020 |
0.1317615093 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 10 |
Hb_000069_410 |
0.1366344297 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 11 |
Hb_111198_020 |
0.138523917 |
- |
- |
hypothetical protein JCGZ_06907 [Jatropha curcas] |
| 12 |
Hb_011512_090 |
0.1398169517 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 13 |
Hb_002827_020 |
0.140398853 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 14 |
Hb_000377_090 |
0.1409012883 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
acetyl-CoA C-acetyltransferase [Hevea brasiliensis] |
| 15 |
Hb_000976_040 |
0.1417581068 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 16 |
Hb_003927_010 |
0.1426025867 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_005143_030 |
0.1438341117 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 18 |
Hb_000679_330 |
0.1443202245 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 19 |
Hb_008494_010 |
0.1450710225 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 20 |
Hb_010487_020 |
0.1453816015 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |