| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
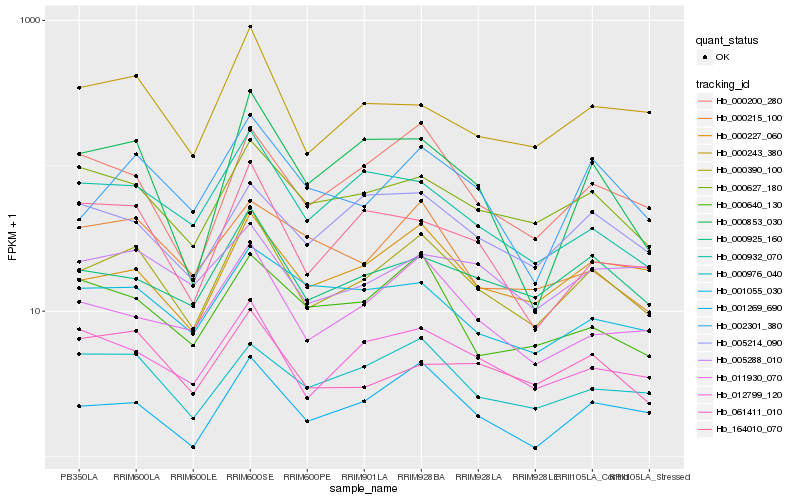
Hb_000390_100 |
0.0 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 2 |
Hb_000227_060 |
0.1141407342 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 3 |
Hb_000925_160 |
0.116124933 |
- |
- |
PREDICTED: digalactosyldiacylglycerol synthase 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000627_180 |
0.1168558036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_012799_120 |
0.1185804704 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002301_380 |
0.1202838296 |
- |
- |
PREDICTED: VQ motif-containing protein 4 [Jatropha curcas] |
| 7 |
Hb_000200_280 |
0.1215180117 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 8 |
Hb_011930_070 |
0.1267194502 |
- |
- |
PREDICTED: probable AMP deaminase [Jatropha curcas] |
| 9 |
Hb_000243_380 |
0.1270783878 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |
| 10 |
Hb_000976_040 |
0.127375899 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 11 |
Hb_000932_070 |
0.1369700187 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 12 |
Hb_001269_690 |
0.1372824877 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_164010_070 |
0.1383790781 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 14 |
Hb_005214_090 |
0.1392290927 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 15 |
Hb_061411_010 |
0.1398226087 |
- |
- |
PREDICTED: ATPase WRNIP1 [Jatropha curcas] |
| 16 |
Hb_000853_030 |
0.1416627289 |
- |
- |
sucrose synthase 2 [Hevea brasiliensis] |
| 17 |
Hb_000640_130 |
0.1422446914 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_001055_030 |
0.1431115964 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000215_100 |
0.1439691173 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005288_010 |
0.1442594048 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |