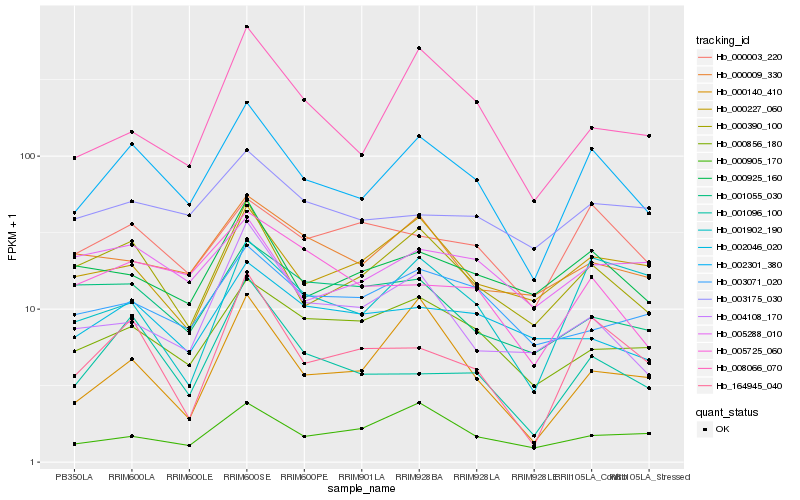
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002301_380 |
0.0 |
- |
- |
PREDICTED: VQ motif-containing protein 4 [Jatropha curcas] |
| 2 |
Hb_000390_100 |
0.1202838296 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 3 |
Hb_000856_180 |
0.1450879717 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 4 |
Hb_164945_040 |
0.1492874084 |
- |
- |
PREDICTED: uncharacterized protein LOC105628261 [Jatropha curcas] |
| 5 |
Hb_000003_220 |
0.1519789267 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 6 |
Hb_000925_160 |
0.1537153477 |
- |
- |
PREDICTED: digalactosyldiacylglycerol synthase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000227_060 |
0.1558716664 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 8 |
Hb_001096_100 |
0.1584778195 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005725_060 |
0.1600946319 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |
| 10 |
Hb_000009_330 |
0.160494662 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 11 |
Hb_008066_070 |
0.1616006712 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 12 |
Hb_000905_170 |
0.1636620003 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 13 |
Hb_001902_190 |
0.1651163925 |
- |
- |
PREDICTED: oxalate--CoA ligase [Jatropha curcas] |
| 14 |
Hb_003071_020 |
0.1652222564 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 15 |
Hb_003175_030 |
0.166135066 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 16 |
Hb_002046_020 |
0.1664277558 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 17 |
Hb_000140_410 |
0.1679712741 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005288_010 |
0.168267157 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_001055_030 |
0.1738865276 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004108_170 |
0.174525786 |
- |
- |
conserved hypothetical protein [Ricinus communis] |