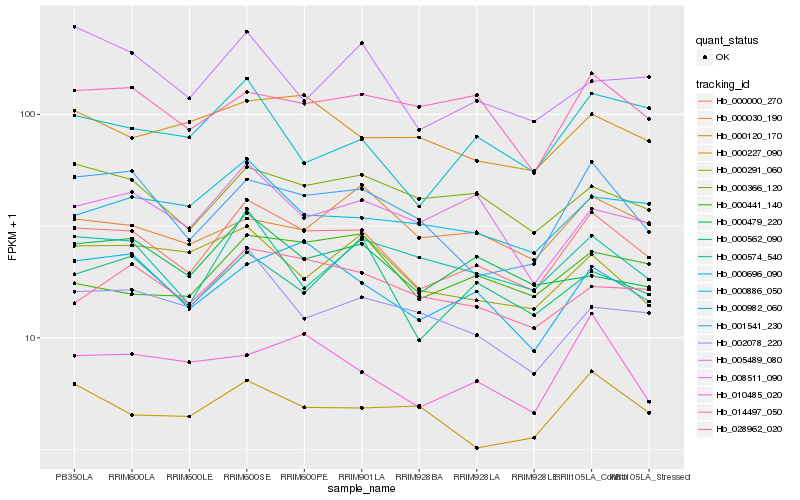
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000000_270 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185 [Jatropha curcas] |
| 2 |
Hb_000366_120 |
0.0768560139 |
- |
- |
PREDICTED: chaperone protein dnaJ 49-like [Jatropha curcas] |
| 3 |
Hb_000982_060 |
0.0790442509 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 4 |
Hb_000562_090 |
0.0823611415 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 6 [Jatropha curcas] |
| 5 |
Hb_001541_230 |
0.084103267 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_028962_020 |
0.0841594246 |
- |
- |
hypothetical protein POPTR_0016s00660g [Populus trichocarpa] |
| 7 |
Hb_000886_050 |
0.0848694424 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 8 |
Hb_000479_220 |
0.0856732926 |
- |
- |
PREDICTED: uncharacterized protein LOC105644125 [Jatropha curcas] |
| 9 |
Hb_000574_540 |
0.0867213983 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_014497_050 |
0.0871461884 |
transcription factor |
TF Family: bZIP |
hypothetical protein JCGZ_08021 [Jatropha curcas] |
| 11 |
Hb_000227_090 |
0.0885281633 |
- |
- |
PREDICTED: NEDD8-activating enzyme E1 catalytic subunit [Jatropha curcas] |
| 12 |
Hb_008511_090 |
0.0894290129 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 13 |
Hb_000291_060 |
0.09029964 |
transcription factor |
TF Family: C2H2 |
PREDICTED: MYST-like histone acetyltransferase 2 [Jatropha curcas] |
| 14 |
Hb_000441_140 |
0.0910796394 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-7-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000120_170 |
0.0921953269 |
- |
- |
selenoprotein [Populus trichocarpa] |
| 16 |
Hb_002078_220 |
0.0923460313 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000030_190 |
0.0931084687 |
- |
- |
PREDICTED: TATA-box-binding protein-like isoform X1 [Citrus sinensis] |
| 18 |
Hb_000696_090 |
0.0956615905 |
- |
- |
PREDICTED: uncharacterized protein LOC105647093 [Jatropha curcas] |
| 19 |
Hb_005489_080 |
0.0959833104 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_010485_020 |
0.0961188334 |
- |
- |
PREDICTED: thaumatin-like protein 1b isoform X1 [Jatropha curcas] |