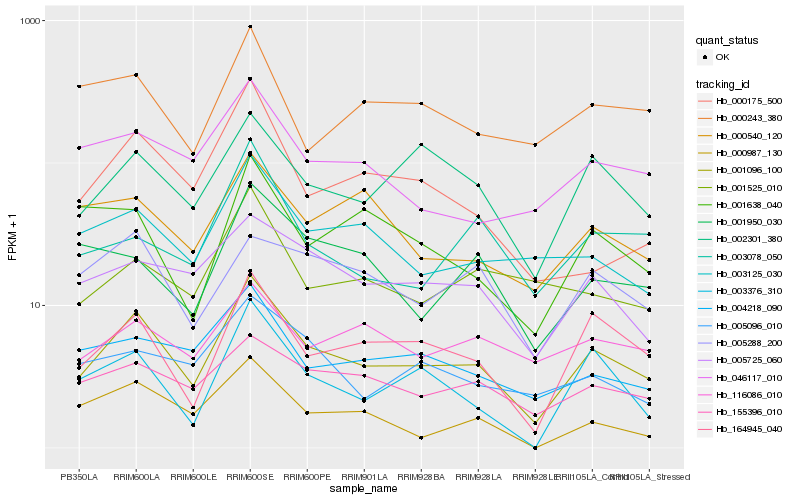
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001096_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_164945_040 |
0.1295559423 |
- |
- |
PREDICTED: uncharacterized protein LOC105628261 [Jatropha curcas] |
| 3 |
Hb_002301_380 |
0.1584778195 |
- |
- |
PREDICTED: VQ motif-containing protein 4 [Jatropha curcas] |
| 4 |
Hb_155396_010 |
0.161464409 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 5 |
Hb_004218_090 |
0.1669537012 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_003125_030 |
0.167421677 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 7 |
Hb_005725_060 |
0.1674405217 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |
| 8 |
Hb_003376_310 |
0.1691853685 |
- |
- |
hypothetical protein JCGZ_16500 [Jatropha curcas] |
| 9 |
Hb_046117_010 |
0.1734650753 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 3 [Jatropha curcas] |
| 10 |
Hb_000175_500 |
0.1764847533 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |
| 11 |
Hb_005096_010 |
0.1778939265 |
- |
- |
PREDICTED: uncharacterized protein LOC105641403 [Jatropha curcas] |
| 12 |
Hb_116086_010 |
0.1790582235 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 13 |
Hb_001525_010 |
0.179762317 |
- |
- |
PREDICTED: uncharacterized protein LOC105638298 [Jatropha curcas] |
| 14 |
Hb_000987_130 |
0.1799910698 |
- |
- |
- |
| 15 |
Hb_001638_040 |
0.1805406276 |
- |
- |
Nucleobase-ascorbate transporter 3 [Glycine soja] |
| 16 |
Hb_000540_120 |
0.1813674207 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 17 |
Hb_003078_050 |
0.1834810582 |
- |
- |
PREDICTED: serine/threonine-protein kinase OXI1 [Jatropha curcas] |
| 18 |
Hb_000243_380 |
0.1835468442 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |
| 19 |
Hb_001950_030 |
0.1841870926 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 20 |
Hb_005288_200 |
0.1844796482 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |