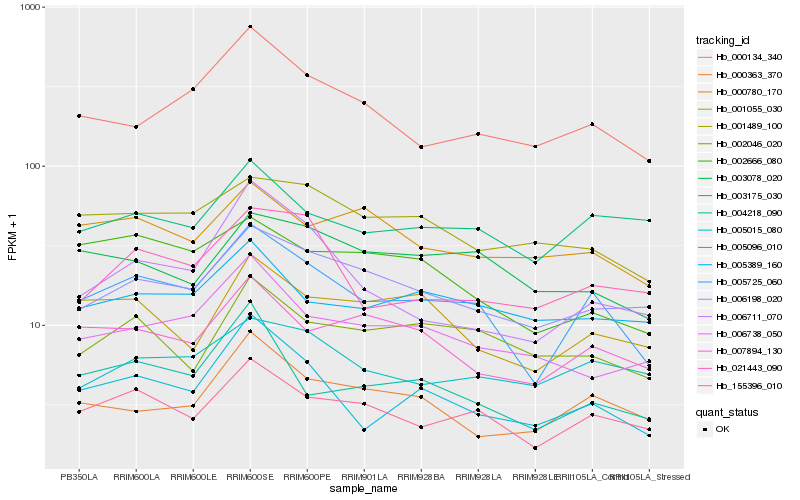
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005725_060 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |
| 2 |
Hb_155396_010 |
0.0975702161 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 3 |
Hb_005096_010 |
0.1183681306 |
- |
- |
PREDICTED: uncharacterized protein LOC105641403 [Jatropha curcas] |
| 4 |
Hb_006198_020 |
0.1263126772 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 5 |
Hb_000134_340 |
0.1281781839 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 5 [Jatropha curcas] |
| 6 |
Hb_007894_130 |
0.131621694 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |
| 7 |
Hb_004218_090 |
0.1379314256 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000363_370 |
0.1382647279 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 [Jatropha curcas] |
| 9 |
Hb_001055_030 |
0.1411273552 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002666_080 |
0.1420222144 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_001489_100 |
0.142845422 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 12 |
Hb_003175_030 |
0.1430307618 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 13 |
Hb_021443_090 |
0.1450238771 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 14 |
Hb_005015_080 |
0.1450901541 |
- |
- |
PREDICTED: shikimate kinase 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_006738_050 |
0.1452071131 |
- |
- |
PREDICTED: uncharacterized protein LOC105634992 [Jatropha curcas] |
| 16 |
Hb_003078_020 |
0.1461505231 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 17 |
Hb_002046_020 |
0.1467574864 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 18 |
Hb_005389_160 |
0.1477867314 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 19 |
Hb_000780_170 |
0.1489693063 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 20 |
Hb_006711_070 |
0.1489842926 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |