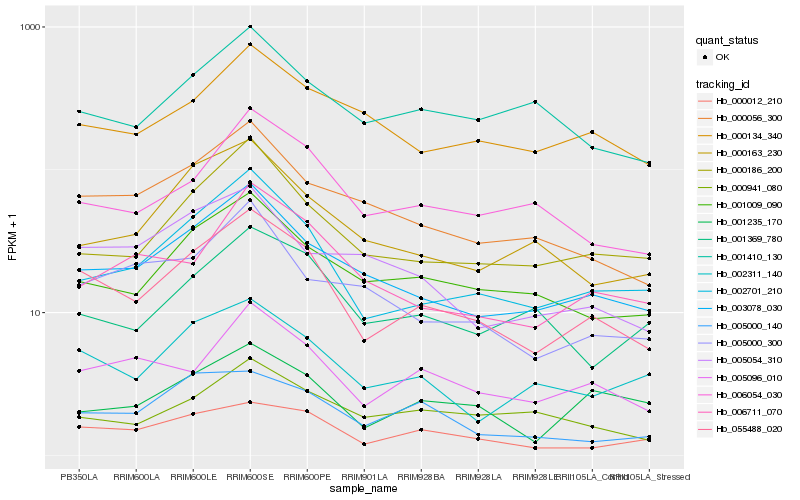
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_055488_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641270 [Jatropha curcas] |
| 2 |
Hb_002701_210 |
0.1142192942 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 3 |
Hb_003078_030 |
0.1146066338 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 4 |
Hb_000012_210 |
0.1192656639 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001009_090 |
0.1310544588 |
- |
- |
CDK protein [Hevea brasiliensis] |
| 6 |
Hb_005096_010 |
0.1322729133 |
- |
- |
PREDICTED: uncharacterized protein LOC105641403 [Jatropha curcas] |
| 7 |
Hb_000056_300 |
0.1327285971 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000186_200 |
0.1331745792 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 9 |
Hb_002311_140 |
0.1333481896 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 15-like [Jatropha curcas] |
| 10 |
Hb_006054_030 |
0.1374776348 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 11 |
Hb_000134_340 |
0.1487653183 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 5 [Jatropha curcas] |
| 12 |
Hb_000163_230 |
0.1512955514 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 13 |
Hb_005000_140 |
0.151532245 |
- |
- |
PREDICTED: uncharacterized protein LOC105637876 isoform X1 [Jatropha curcas] |
| 14 |
Hb_006711_070 |
0.1533585216 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 15 |
Hb_001410_130 |
0.1539100591 |
- |
- |
hypothetical protein POPTR_0001s10880g [Populus trichocarpa] |
| 16 |
Hb_001235_170 |
0.1557053061 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 17 |
Hb_001369_780 |
0.1567961109 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000941_080 |
0.1579472016 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 7-like isoform X3 [Populus euphratica] |
| 19 |
Hb_005054_310 |
0.1582005104 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 20 |
Hb_005000_300 |
0.1598950373 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |