| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
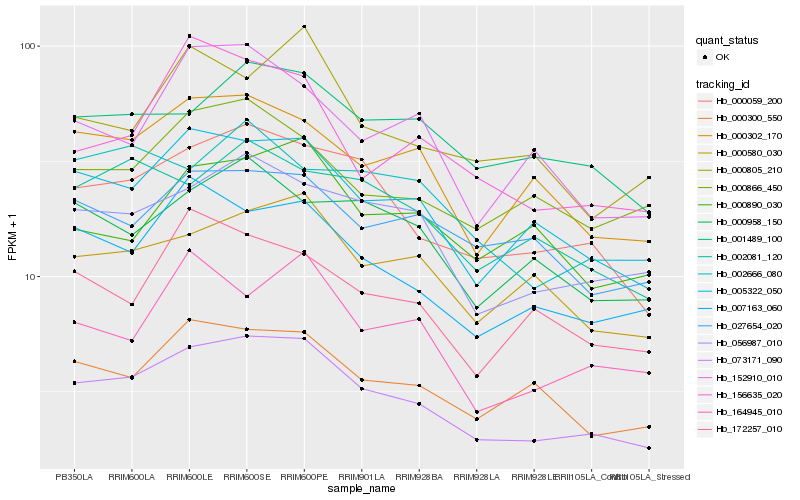
Hb_073171_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |
| 2 |
Hb_000059_200 |
0.0736540998 |
- |
- |
PREDICTED: WEB family protein At2g38370-like [Jatropha curcas] |
| 3 |
Hb_005322_050 |
0.0776746621 |
- |
- |
hypothetical protein POPTR_0009s02400g [Populus trichocarpa] |
| 4 |
Hb_056987_010 |
0.0867833828 |
- |
- |
DNA-damage-repair/toleration protein DRT111, chloroplastic [Glycine soja] |
| 5 |
Hb_007163_060 |
0.0869855095 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 6 |
Hb_000300_550 |
0.0943522122 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 7 |
Hb_000302_170 |
0.0946028716 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |
| 8 |
Hb_000580_030 |
0.0959283719 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 9 |
Hb_001489_100 |
0.106188352 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 10 |
Hb_000958_150 |
0.1076806828 |
- |
- |
Actin, putative [Ricinus communis] |
| 11 |
Hb_002081_120 |
0.107737983 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 12 |
Hb_000866_450 |
0.1093219156 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 13 |
Hb_172257_010 |
0.1103044098 |
transcription factor |
TF Family: HMG |
high mobility group family protein [Populus trichocarpa] |
| 14 |
Hb_000805_210 |
0.1104902446 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 15 |
Hb_164945_010 |
0.1110988984 |
- |
- |
PREDICTED: DNA primase small subunit [Jatropha curcas] |
| 16 |
Hb_152910_010 |
0.1111460617 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 17 |
Hb_027654_020 |
0.1115683745 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Jatropha curcas] |
| 18 |
Hb_156635_020 |
0.1119208355 |
- |
- |
PREDICTED: L-lactate dehydrogenase B [Jatropha curcas] |
| 19 |
Hb_000890_030 |
0.1148150239 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 20 |
Hb_002666_080 |
0.117098675 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |