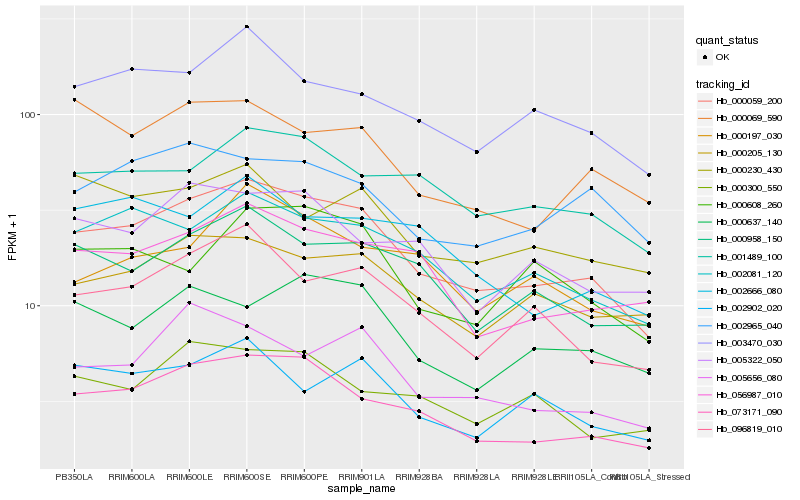
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000059_200 |
0.0 |
- |
- |
PREDICTED: WEB family protein At2g38370-like [Jatropha curcas] |
| 2 |
Hb_073171_090 |
0.0736540998 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |
| 3 |
Hb_002081_120 |
0.0907434914 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 4 |
Hb_096819_010 |
0.0917188502 |
- |
- |
PREDICTED: GRIP and coiled-coil domain-containing protein 2 isoform X4 [Jatropha curcas] |
| 5 |
Hb_000958_150 |
0.0965990404 |
- |
- |
Actin, putative [Ricinus communis] |
| 6 |
Hb_003470_030 |
0.0984750676 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |
| 7 |
Hb_000205_130 |
0.1000340399 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 8 |
Hb_056987_010 |
0.1015892822 |
- |
- |
DNA-damage-repair/toleration protein DRT111, chloroplastic [Glycine soja] |
| 9 |
Hb_001489_100 |
0.1038650158 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 10 |
Hb_005656_080 |
0.1061523975 |
- |
- |
PREDICTED: RNA-binding KH domain-containing protein PEPPER-like [Jatropha curcas] |
| 11 |
Hb_005322_050 |
0.1101868758 |
- |
- |
hypothetical protein POPTR_0009s02400g [Populus trichocarpa] |
| 12 |
Hb_000637_140 |
0.1126302954 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 13 |
Hb_000300_550 |
0.1139740893 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 14 |
Hb_002902_020 |
0.1144421355 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 15 |
Hb_002965_040 |
0.1144627457 |
- |
- |
drought-responsive family protein [Populus trichocarpa] |
| 16 |
Hb_000197_030 |
0.115240812 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 17 |
Hb_000230_430 |
0.1159411752 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_000608_260 |
0.1160870235 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 19 |
Hb_002666_080 |
0.1163246259 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_000069_590 |
0.1166155761 |
- |
- |
auxin-responsive GH3 family protein [Hevea brasiliensis] |