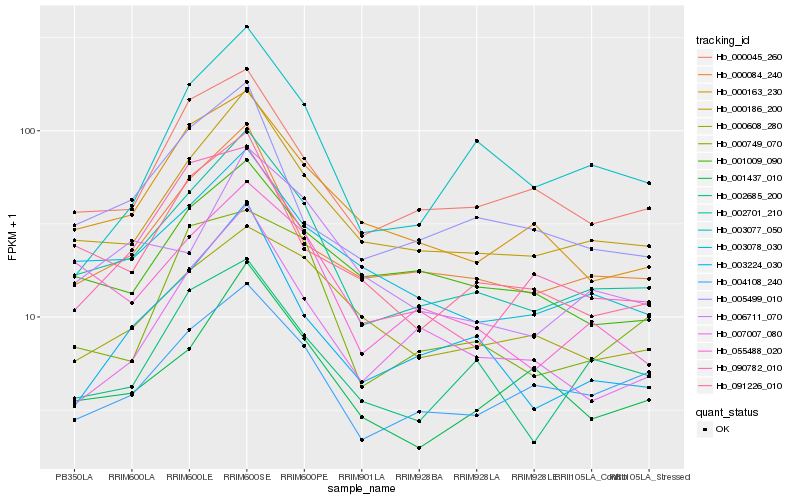
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002701_210 |
0.0 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 2 |
Hb_000186_200 |
0.0723956235 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 3 |
Hb_003078_030 |
0.103241223 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 4 |
Hb_000084_240 |
0.1033939111 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 5 |
Hb_000163_230 |
0.1126858368 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 6 |
Hb_000045_260 |
0.1128887991 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_004108_240 |
0.1133500241 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 8 |
Hb_055488_020 |
0.1142192942 |
- |
- |
PREDICTED: uncharacterized protein LOC105641270 [Jatropha curcas] |
| 9 |
Hb_001437_010 |
0.1213898689 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g33260 [Jatropha curcas] |
| 10 |
Hb_091226_010 |
0.1221616276 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA isoform X1 [Jatropha curcas] |
| 11 |
Hb_003224_030 |
0.1238191895 |
- |
- |
hypothetical protein POPTR_0077s002402g, partial [Populus trichocarpa] |
| 12 |
Hb_002685_200 |
0.1252792393 |
- |
- |
PREDICTED: uncharacterized protein LOC105632583 [Jatropha curcas] |
| 13 |
Hb_001009_090 |
0.1284398895 |
- |
- |
CDK protein [Hevea brasiliensis] |
| 14 |
Hb_007007_080 |
0.1293267041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000749_070 |
0.1294016514 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 16 |
Hb_090782_010 |
0.1310268399 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 17 |
Hb_005499_010 |
0.1317122673 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 18 |
Hb_006711_070 |
0.134761989 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 19 |
Hb_003077_050 |
0.137689593 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 20 |
Hb_000608_280 |
0.1404578699 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |