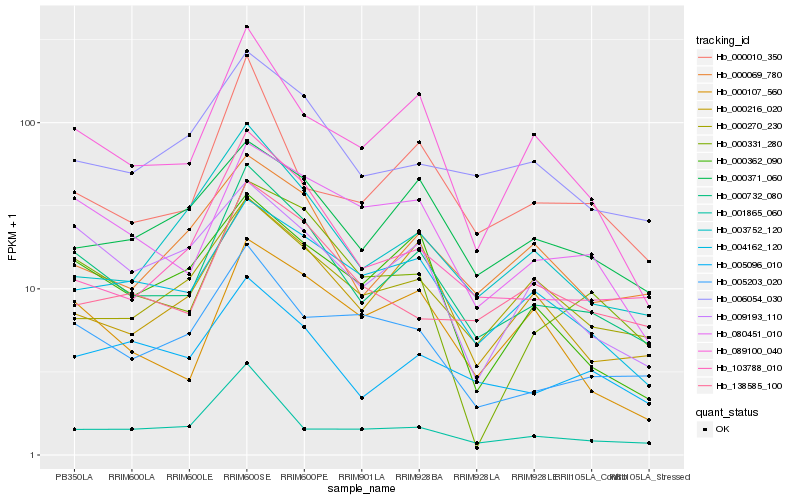
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000732_080 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g45440 [Jatropha curcas] |
| 2 |
Hb_103788_010 |
0.1267059968 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 3 |
Hb_089100_040 |
0.1281615001 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |
| 4 |
Hb_004162_120 |
0.1310430353 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 41 homolog [Populus euphratica] |
| 5 |
Hb_006054_030 |
0.13156105 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 6 |
Hb_080451_010 |
0.1323679072 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680 [Jatropha curcas] |
| 7 |
Hb_005096_010 |
0.1345588532 |
- |
- |
PREDICTED: uncharacterized protein LOC105641403 [Jatropha curcas] |
| 8 |
Hb_000216_020 |
0.1346098154 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 9 |
Hb_000069_780 |
0.1378642112 |
- |
- |
atpob1, putative [Ricinus communis] |
| 10 |
Hb_009193_110 |
0.1437554611 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000331_280 |
0.1442025388 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 12 |
Hb_005203_020 |
0.1442115435 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 13 |
Hb_000270_230 |
0.1461154595 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 14 |
Hb_000371_060 |
0.1474449393 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 15 |
Hb_003752_120 |
0.1478159411 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000107_560 |
0.1516514914 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |
| 17 |
Hb_000010_350 |
0.1518464579 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 18 |
Hb_138585_100 |
0.1529340297 |
- |
- |
PREDICTED: uncharacterized protein LOC105648740 [Jatropha curcas] |
| 19 |
Hb_001865_060 |
0.1529425839 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 20 |
Hb_000362_090 |
0.1533243393 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |