| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
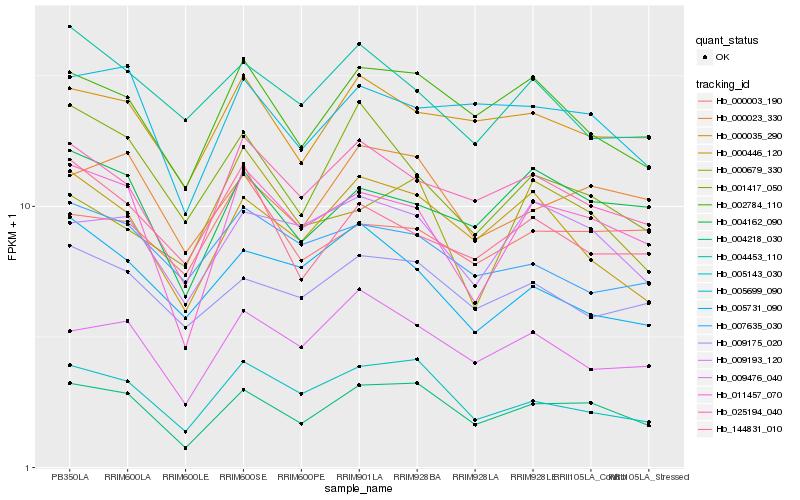
Hb_011457_070 |
0.0 |
- |
- |
5-methyltetrahydrofolate:homocysteine methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_004218_030 |
0.0851129622 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004162_090 |
0.0912950342 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 4 |
Hb_025194_040 |
0.0974612385 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X2 [Jatropha curcas] |
| 5 |
Hb_009193_120 |
0.1058745413 |
desease resistance |
Gene Name: AAA_22 |
PREDICTED: putative ATP-dependent RNA helicase DHX33 [Jatropha curcas] |
| 6 |
Hb_005143_030 |
0.1105531024 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 7 |
Hb_009476_040 |
0.1145440249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001417_050 |
0.1194465879 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 9 |
Hb_144831_010 |
0.1204700747 |
transcription factor |
TF Family: MYB |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_005699_090 |
0.1207777928 |
- |
- |
PREDICTED: uncharacterized protein LOC105639563 [Jatropha curcas] |
| 11 |
Hb_002784_110 |
0.1211318196 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 12 |
Hb_000446_120 |
0.1218128448 |
- |
- |
PREDICTED: uncharacterized transmembrane protein DDB_G0289901 [Jatropha curcas] |
| 13 |
Hb_000003_190 |
0.122519829 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like isoform X1 [Nelumbo nucifera] |
| 14 |
Hb_005731_090 |
0.1227215167 |
- |
- |
PREDICTED: uncharacterized protein LOC105630164 [Jatropha curcas] |
| 15 |
Hb_000035_290 |
0.124246422 |
- |
- |
RNA recognition motif-containing family protein [Populus trichocarpa] |
| 16 |
Hb_000679_330 |
0.1244689292 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 17 |
Hb_000023_330 |
0.1248038136 |
- |
- |
RNA recognition motif-containing family protein [Populus trichocarpa] |
| 18 |
Hb_004453_110 |
0.1257597804 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 19 |
Hb_009175_020 |
0.126022879 |
- |
- |
PREDICTED: uncharacterized protein LOC101303140 [Fragaria vesca subsp. vesca] |
| 20 |
Hb_007635_030 |
0.1263402133 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |