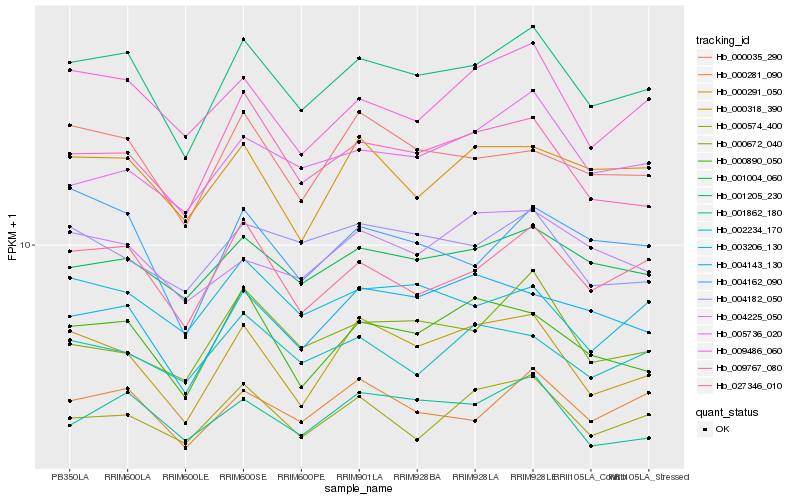
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001205_230 |
0.0 |
- |
- |
PREDICTED: acidic leucine-rich nuclear phosphoprotein 32-related protein [Jatropha curcas] |
| 2 |
Hb_027346_010 |
0.0545915729 |
- |
- |
- |
| 3 |
Hb_001004_060 |
0.0605353945 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 4 |
Hb_000281_090 |
0.0667754765 |
transcription factor |
TF Family: SNF2 |
hypothetical protein PRUPE_ppa000063mg [Prunus persica] |
| 5 |
Hb_009767_080 |
0.0688121615 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 6 |
Hb_009486_060 |
0.069521196 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 7 |
Hb_000035_290 |
0.0743157891 |
- |
- |
RNA recognition motif-containing family protein [Populus trichocarpa] |
| 8 |
Hb_000890_050 |
0.0761085412 |
- |
- |
PREDICTED: uncharacterized protein LOC105631800 [Jatropha curcas] |
| 9 |
Hb_004182_050 |
0.076770287 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001862_180 |
0.0775166117 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein ELI1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003206_130 |
0.0776831511 |
- |
- |
PREDICTED: protein NRDE2 homolog [Jatropha curcas] |
| 12 |
Hb_000318_390 |
0.077748127 |
- |
- |
PREDICTED: uncharacterized protein LOC105631693 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000291_050 |
0.0792926416 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105638874 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002234_170 |
0.0796307175 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_004143_130 |
0.0798884817 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsC [Jatropha curcas] |
| 16 |
Hb_000574_400 |
0.0806935068 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g15930 [Jatropha curcas] |
| 17 |
Hb_004162_090 |
0.0815118251 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 18 |
Hb_005736_020 |
0.0818447852 |
- |
- |
hypothetical protein POPTR_0005s20940g [Populus trichocarpa] |
| 19 |
Hb_000672_040 |
0.0824385873 |
- |
- |
PREDICTED: ENHANCER OF AG-4 protein 2 [Jatropha curcas] |
| 20 |
Hb_004225_050 |
0.08274 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |